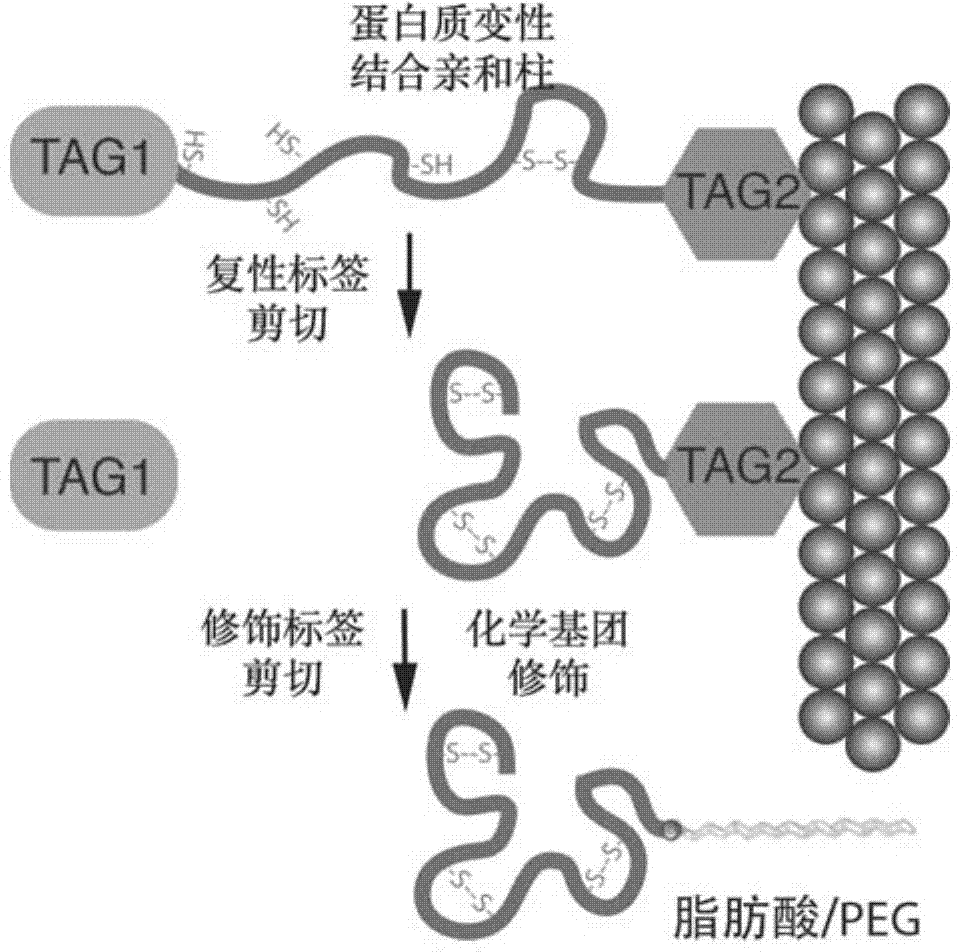
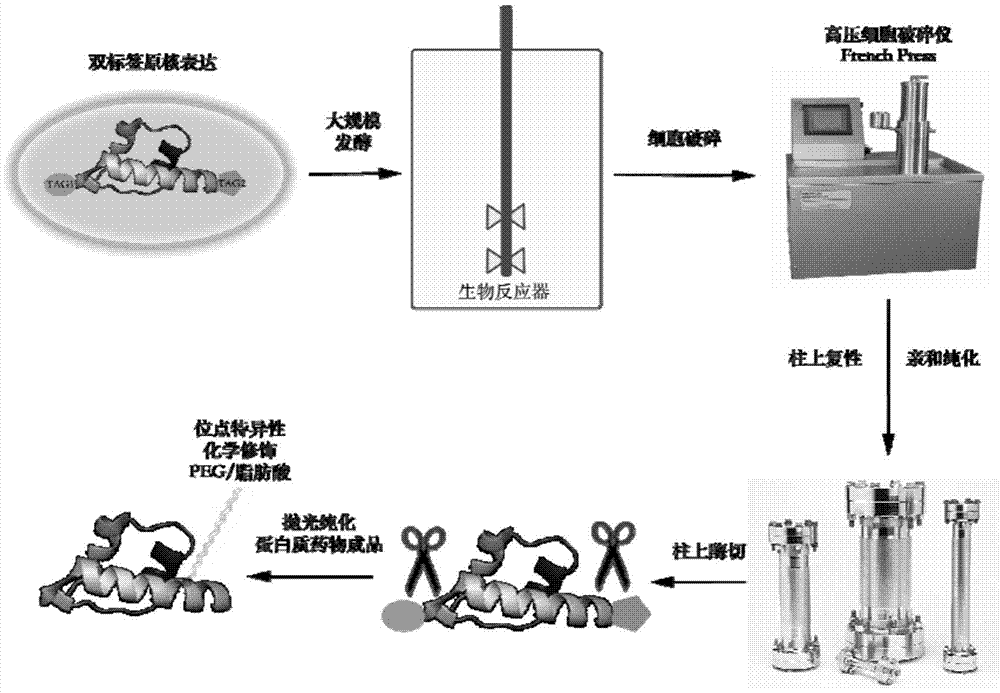
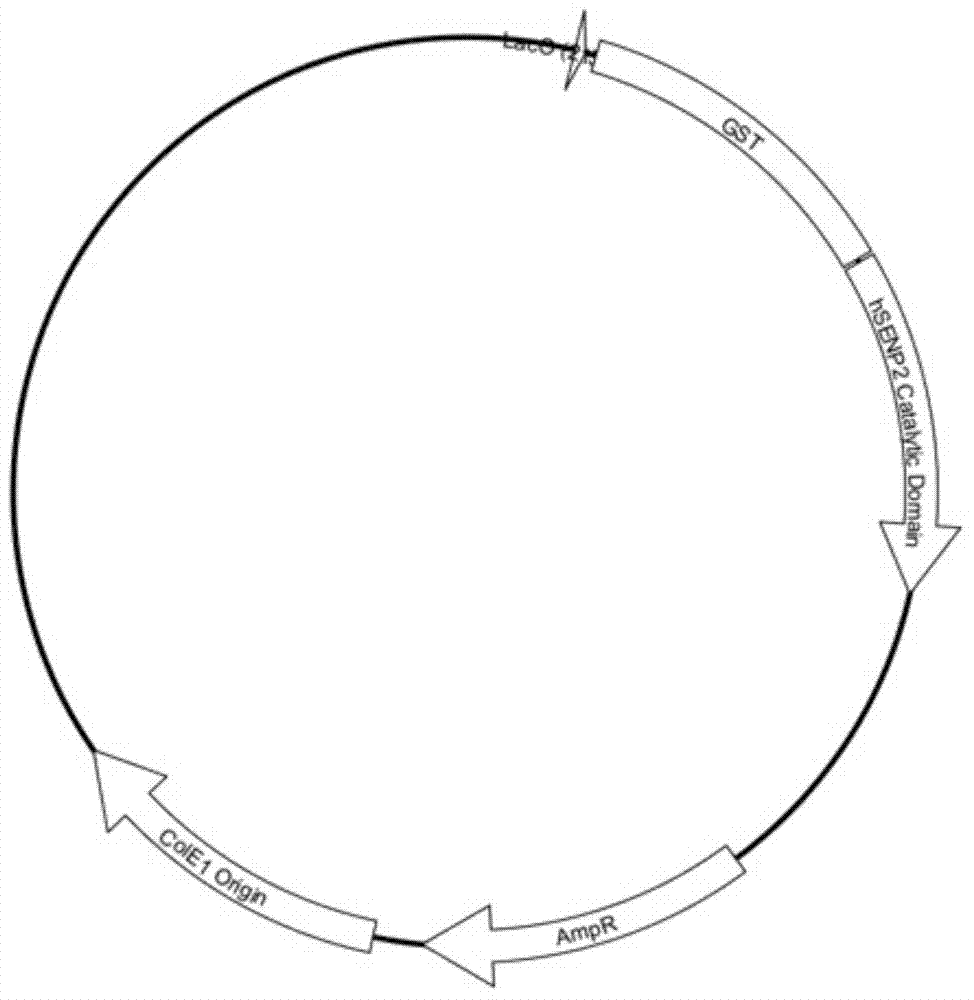
Double-label vector for expression, refolding and modification of long-acting protein medicament and method
A protein and double tag technology, applied in the field of genetic engineering, can solve the problems of efficient and difficult to take into account of proteins, and achieve the effect of promoting construction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0081] Embodiment 1: Construction of pSumo-Intein plasmid vector
[0082] (1) Obtain the humanized Sumo3 protein sequence (P55854) through the Uniprot database, and then use the protein sequence reverse translation and codon optimization service (http: / / genomes.urv.es / OPTIMIZER / ) to obtain the Sumo3 DNA sequence ( SEQ ID No. 1). On this basis, its 7-276 bp was taken as the sequence of sumo3tag (SEQ ID No.2), and this sequence also contained a little 6×His tag sequence to facilitate IMAC affinity denaturation and purification later.
[0083] (2) Next, design segmented primers (Gene Design, http: / / 54.235.254.95 / gd / ) for gene synthesis on the optimized DNA sequence:
[0084] The synthesis reaction is accomplished by Assembly PCR reaction (Marchand, J.A., and Peccoud, J. (2012). Building block synthesis using the polymerase chain assembly method. Methods Mol. Biol. 852, 3–10.). The sequences OPM and TPM of the segmented primer are shown in SEQ ID NO.6-15. All designed Overlap p...
Embodiment 2
[0088] Embodiment 2: Preparation of human source SENP2 catalytic domain protein
[0089] (1) Obtain the human SENP2 protein sequence (Q9POU3) through the Uniprot database, and then use the protein sequence reverse translation and codon optimization service (http: / / genomes.urv.es / OPTIMIZER / ) to obtain the SENP2 catalytic domain (362-589aa ) DNA sequence (SEQ ID No.3);
[0090] (2) Next, design segmented primers (Gene Design, http: / / 54.235.254.95 / gd / ) for gene synthesis on the optimized DNA sequence:
[0091] The sequences OPM and TPM of the segmented primer are shown in SEQ ID NO.26-43. The synthesis reaction is accomplished by Assembly PCR reaction (Marchand, J.A., and Peccoud, J. (2012). Building block synthesis using the polymerase chain assembly method. Methods Mol. Biol. 852, 3–10.). All designed Overlap primers (ie segmented primers) were synthesized by Sigma and dissolved in aqueous solution to form a 100uM stock solution. Prepare template primer mix (TPM) (SEQ ID NO....
Embodiment 3
[0099] Example 3: Plasmid Construction of Proinsulin and GLP1 Analogs in pSumo-Intein System
[0100] (1), construction of pSumo-proinsulin-Intein plasmid vector
[0101] a. Obtain the sequence of human Proinsulin protein (P01308) through the Uniprot database, and then obtain the prokaryotic expression mutant through the program. The sequence of the mutant is separated by using KR as the linker sequence between the B chain and A chain sequences of Insulin, Form the structure of B chain-KR-A chain; then use protein sequence reverse translation and codon optimization service (http: / / genomes.urv.es / OPTIMIZER / ) to obtain its DNA sequence (SEQ ID No.4).
[0102] b. Design segmented primers (Gene Design, http: / / 54.235.254.95 / gd / ) for the above-mentioned optimized DNA sequence for gene synthesis:
[0103] The synthesis reaction was accomplished by Assembly PCR (Marchand, J.A., and Peccoud, J. (2012). Building block synthesis using the polymerase chain assembly method. Methods Mol. B...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com