Method for rapidly detecting polymorphism of saccharum sucrose phosphate synthase B (SPSB) gene and application of method
A technology of gene polymorphism, SPSB2177P2, which is applied in biochemical equipment and methods, and microbial determination/inspection, etc., can solve the problems that polymorphism analysis of intron regions has not been reported, and intron regions have not been cloned, etc. , to overcome the effects of high cost, high accuracy and strong primer specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
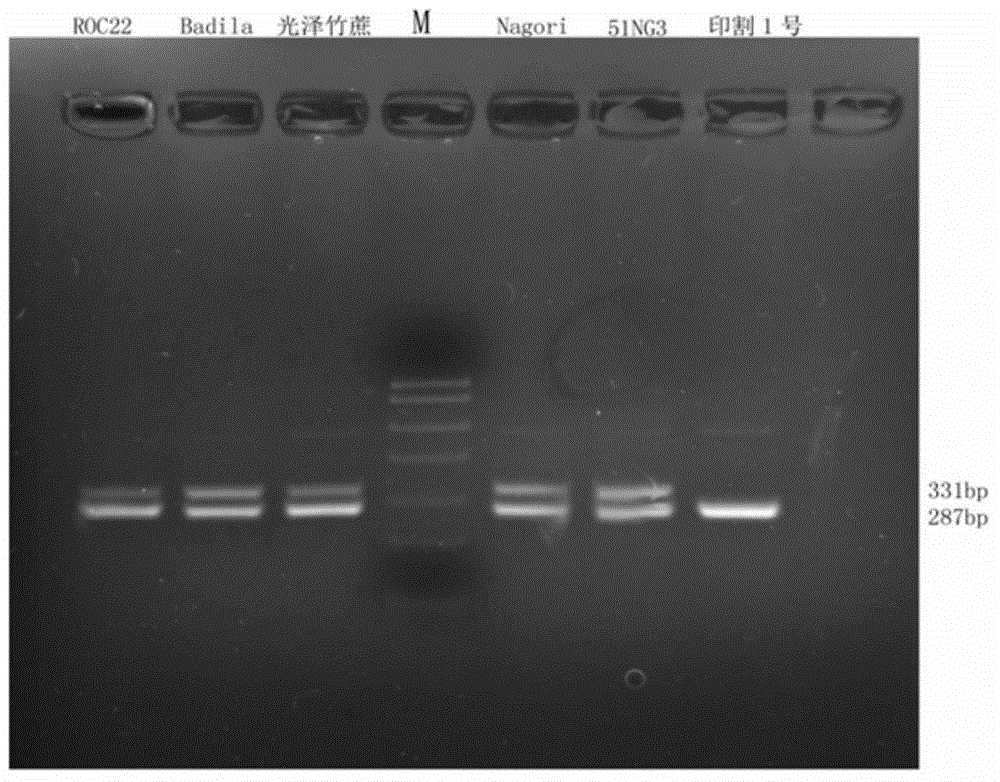
[0047] (1) Cloning and polymorphism detection of partial DNA sequences of SPSB genes in different species of Saccharum.
[0048] 1. Extraction of sample DNA
[0049] The present invention takes sugarcane large-stem wild species 51NG3, Indian species Nagori, sugarcane variety ROC22, tropical species Badila, Chinese species glossy bamboo cane and sugarcane thin-stem raw species Yinke No. 1 as detection objects, and uses EasyPure Plant provided by Quanshijin Company Genomic DNA Kit was used to extract genomic DNA, and finally dissolved in sterilized ultrapure water, and stored at 4°C after detection. The OD value of the DNA samples was measured with a UV photometer. Calculate the DNA content and the ratio of OD260 / OD280. If the ratio of OD260 / OD280 is less than 1.6, it means that the sample contains more protein or phenol, and purification should be performed; if the ratio is greater than 1.8, RNA purification should be considered. After the DNA detection is completed, a certai...
Embodiment 2
[0072] (1) Cloning of partial DNA sequences of SPSB genes of different species of Saccharum and detection of polymorphisms
[0073] 1. Extraction of sample DNA
[0074] In the present invention, the wild sugarcane species 51NG3, the Indian species Nagori, the sugarcane variety ROC22, the tropical species Badila and the Chinese species glossy bamboo cane are used as detection objects, and the Genomic DNA is extracted with the EasyPure Plant Genomic DNA Kit provided by Quanshijin Company. Dissolve in sterilized ultrapure water and store at 4°C after detection. The OD value of the DNA samples was measured with a UV photometer. Calculate the DNA content and the ratio of OD260 / OD280. If the ratio of OD260 / OD280 is less than 1.6, it means that the sample contains more protein or phenol, and purification should be performed; if the ratio is greater than 1.8, RNA purification should be considered. After the DNA detection is completed, a certain amount is taken out and diluted to 40n...
Embodiment 3
[0098] (1) Cloning and polymorphism detection of the partial DNA sequence of the SPSB gene of the wild sugarcane thin-stalked species Yinque 1.
[0099] 1. Extraction of sample DNA
[0100] In the present invention, sugarcane slender stem seed Yincut No. 1 is used as the detection object, the Genomic DNA is extracted with the EasyPure Plant Genomic DNA Kit provided by Quanshijin Company, and finally dissolved in sterilized ultrapure water, and stored at 4°C after detection. The OD value of the DNA samples was measured with a UV photometer. Calculate the DNA content and the ratio of OD260 / OD280. If the ratio of OD260 / OD280 is less than 1.6, it means that the sample contains more protein or phenol, and purification should be performed; if the ratio is greater than 1.8, RNA purification should be considered. After the DNA detection is completed, a certain amount is taken out and diluted to 40ng / μL for use as a PCR template.
[0101] 2. Primer design
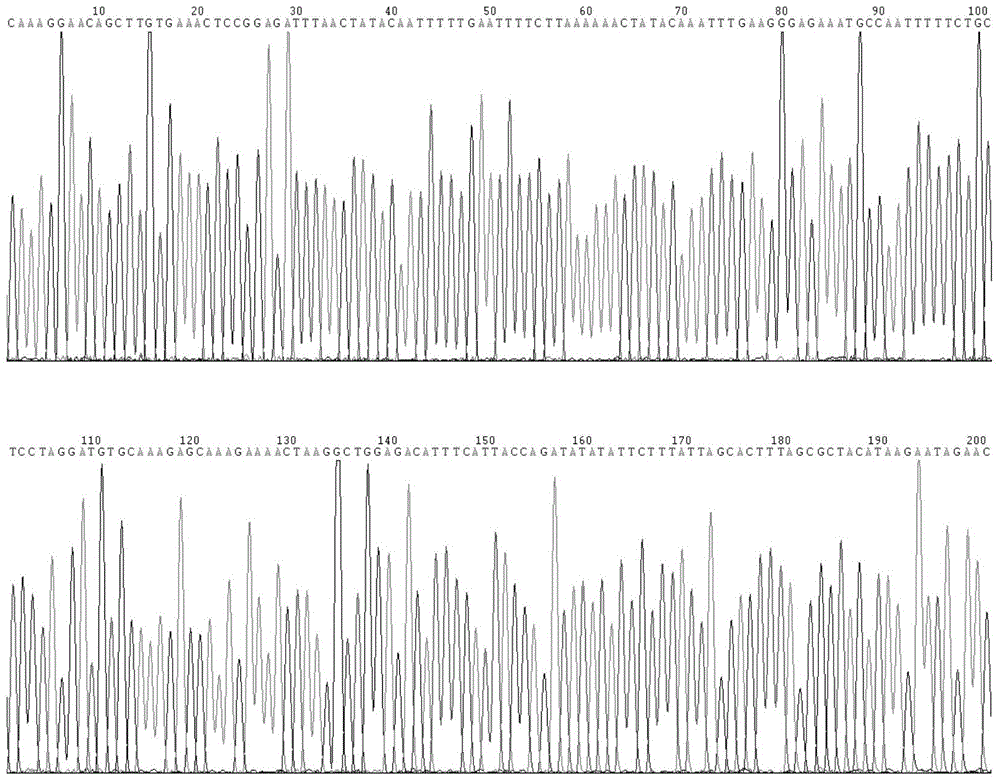
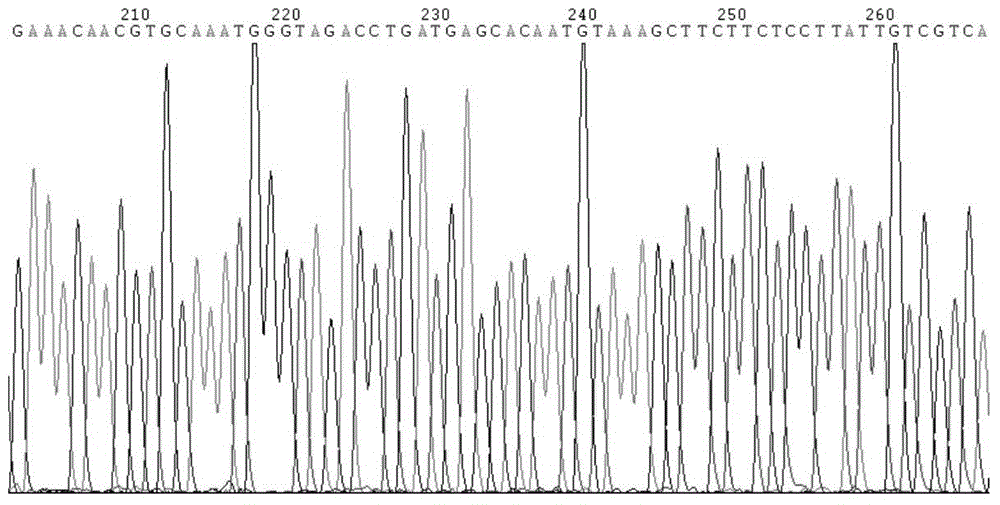
[0102] Obtain the publish...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com