Recombined engineering bacterium for producing UDPG (uridine diphosphate glucose) and application thereof
A technology of recombinant engineering bacteria and engineering bacteria, applied in bacteria, microorganism-based methods, microorganisms, etc., can solve the problems of massive accumulation, affecting the yield of Jinggangmycin A, increasing the cost of Jinggangmycin A, etc., and achieving a fast conversion rate. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
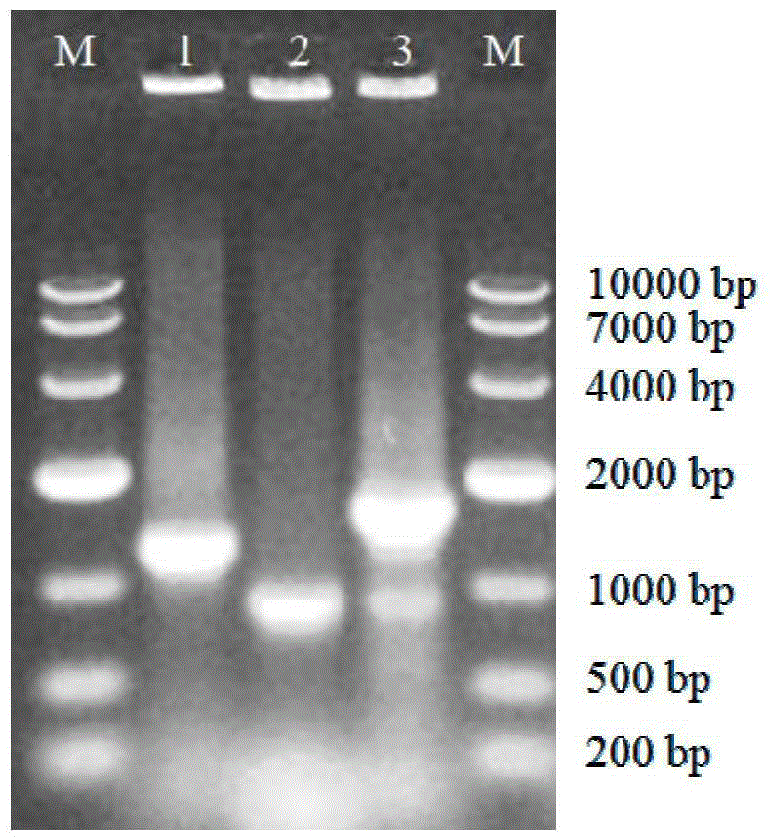
[0030] Example 1 Construction method of Escherichia coli pETDuet-ValG BL21 (DE3) engineering bacteria
[0031] The Streptomyces hygroscopicus genome was obtained by a small extraction method, and then the target gene ValG was introduced into the pETDuet-1 plasmid. The specific method is as follows:
[0032] 1. Mini-extraction of Streptomyces hygroscopicus genome
[0033] Streptomyces hygroscopicus subsp. jinggangensis was inoculated in Bennett's medium, cultured at 28°C and 180rpm for 18h; centrifuged at 12000rpm for 5min, the streptomyces mycelium was collected and washed with 10.3% sucrose aqueous solution for 2 times; with 500 μL, 2mg / mL lysozyme buffer (lysozyme buffer (10mmol / L) composition: 0.3mol / L sucrose, 25mmol / L Tris-HCl (pH8.0), 25mmol / L EDTA (pH8.0 ).) Re-suspend the mycelium, in a water bath at 37°C for 30 minutes until the solution is translucent; add 250 μL of 2% SDS (mass concentration, the solvent is double distilled water), mix and oscillate evenly, and bat...
Embodiment 2
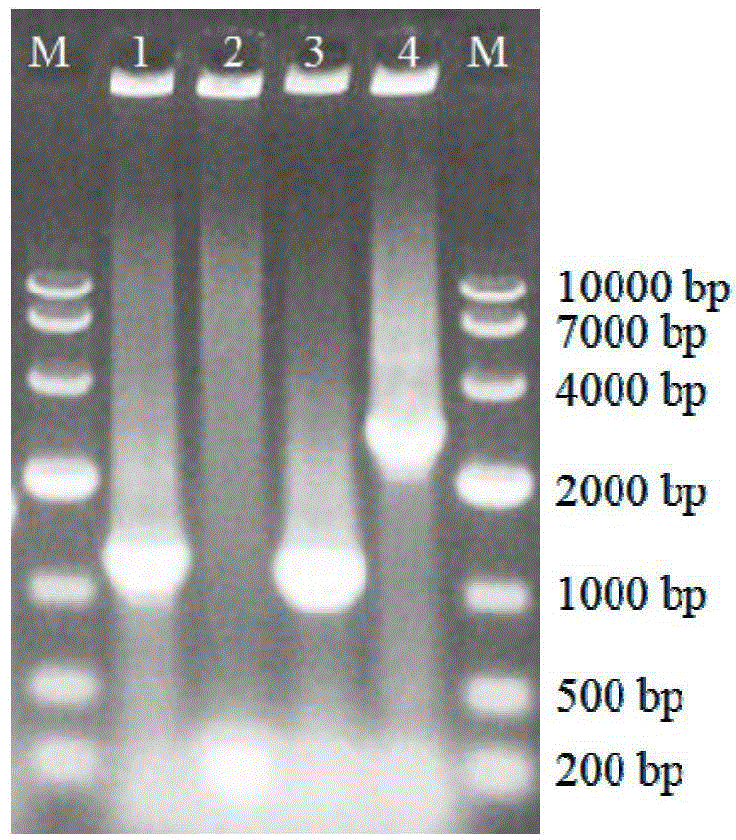
[0041] Example 2 Construction method of Escherichia coli pETDuet-ValG-GalU BL21 (DE3) engineering bacteria
[0042] The E.coli JM109 genome was obtained by the CTAB method, the GalU target gene was amplified, and then the GalU target gene was introduced into the pETDuet-ValG plasmid. The specific method is as follows:
[0043] 1. Mini-extraction of Escherichia coli JM109 genome
[0044] Inoculate E.coli JM109 in LB liquid medium, 37°C, 180rpm, and culture overnight; add the culture solution with a volume concentration of 1% inoculum into fresh LB liquid medium, 37°C, 180rpm, and cultivate for 3h; take 5mL of bacteria solution, centrifuged at 5000rpm for 10min, and collected the bacteria; added 567μL TE buffer solution (pH value 8.0), added 30μL mass concentration 100g / L SDS (solvent double distilled water) and 3μL 20mg / mL proteinase K (solvent double distilled water) mixed Put in a constant temperature water bath at 37°C for 1 hour; add 100 μL 5mol / L NaCl aqueous solution and...
Embodiment 3
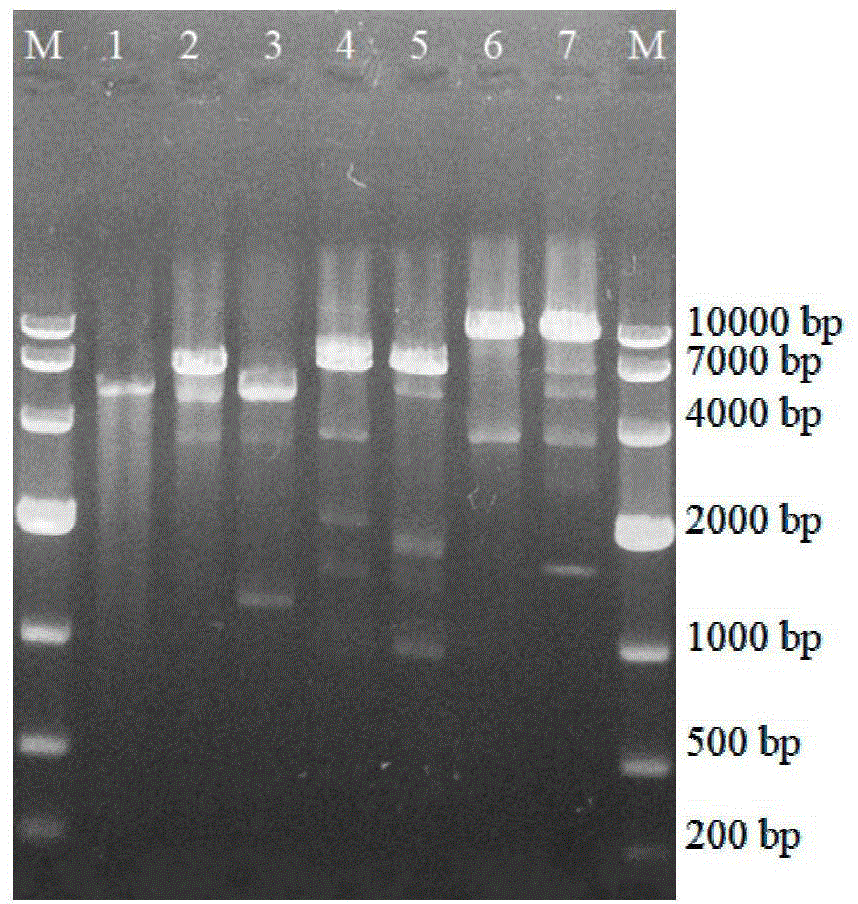
[0056] Example 3 Construction method of Escherichia coli pETDuet-ValG-GalU-Pgm BL21 (DE3) engineering bacteria
[0057] The E.coli JM109 genome was obtained by the CTAB method, the Pgm target gene was obtained by PCR expansion, and then the Pgm target gene was introduced into the pETDuet-ValG-GalU plasmid. The specific method is as follows:
[0058] 1. Amplification of Pgm target gene
[0059] Primers were designed according to the genome sequence of E.coli JM109:
[0060] Pgm-F:5'-TGCA CTGCAGAACGTTGCAGACAAAGGAC-3' (Pst I),
[0061] Pgm-R:5'-ATTT GCGGCCGC TTACGCGTTTTTCAGAAC-3' (Not 1). The Pgm gene was amplified by PCR using the E.coli JM109 genome as the amplification template and Pgm-F / Pgm-R as primers. PCR system: 10× buffer 5.0 μL, d NTPs 4.0 μL, Pgm-F 1.0 μL, Pgm-R 1.0 μL, E.coli JM109 genome template 3.0 μL, TaKaRa Ex Taq HS 0.25
[0062] μL, RNase Free dH 2 O supplemented to 35.75 μL. PCR program: pre-denaturation at 98°C for 2min; denaturation at 98°C for 10s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com