Isobutanol synthetic strain construction method implemented by guiding adjustment of intracellular reducing power based on genomic scale metabolic network model
A genome-scale, metabolic network technology, applied in the field of isobutanol synthesis strain construction based on genome-scale metabolic network model to guide the regulation of intracellular reducing force, can solve the problem of not being able to achieve optimal intracellular reducing force state, strain growth and product Problems such as decreased synthesis ability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0058] Example 1 Construction of vector plasmid pTRCLA and pACYCLA
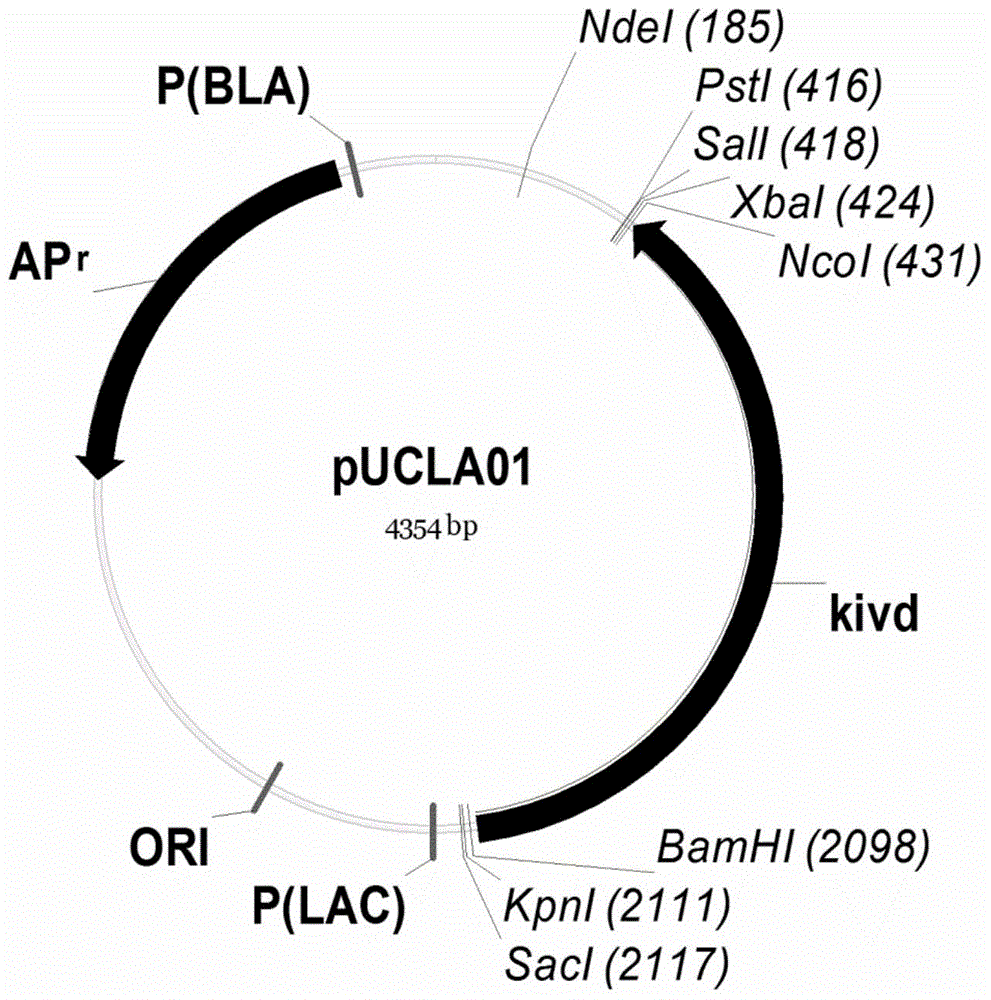
[0059] (1) Using pTRC99a as a template, design primers PLO-F and PLO-R, the primer sequences are SEQ ID NO: 6 and SEQ ID NO: 7, and use PCR to obtain the fragment PLO; using pTRC99a as a template, design primers TB-F and TB-R, the primer sequence is SEQ ID NO: 8 and SEQ ID NO: 9, utilizes PCR to obtain fragment TB; Fragment PLO and TB are digested with KpnI and PstI after double enzymes, ligation obtains plasmid pTRCLA ( Image 6 ).
[0060] Double digestion system: fragment PLO or fragment TB 20 μL, KpnI 1 μL, PstI 1 μL, 10×buffer 1 μL, ddH 2 O 23μL, react at 37°C for 1h.
[0061] Ligation system: TB fragment 7 μL, PLO fragment 1 μL, T4 ligase 0.5 μL, ddH 2 O 1.5 μL, react at 22°C for 1h.
[0062] (2) Using pTRC99a as a template, design primers PT-F and PT-R, the primer sequences are SEQ ID NO: 10 and SEQ ID NO: 11, and use PCR to obtain fragment PT; using pACYC184 as a template, design primers OC-F and ...
Embodiment 2
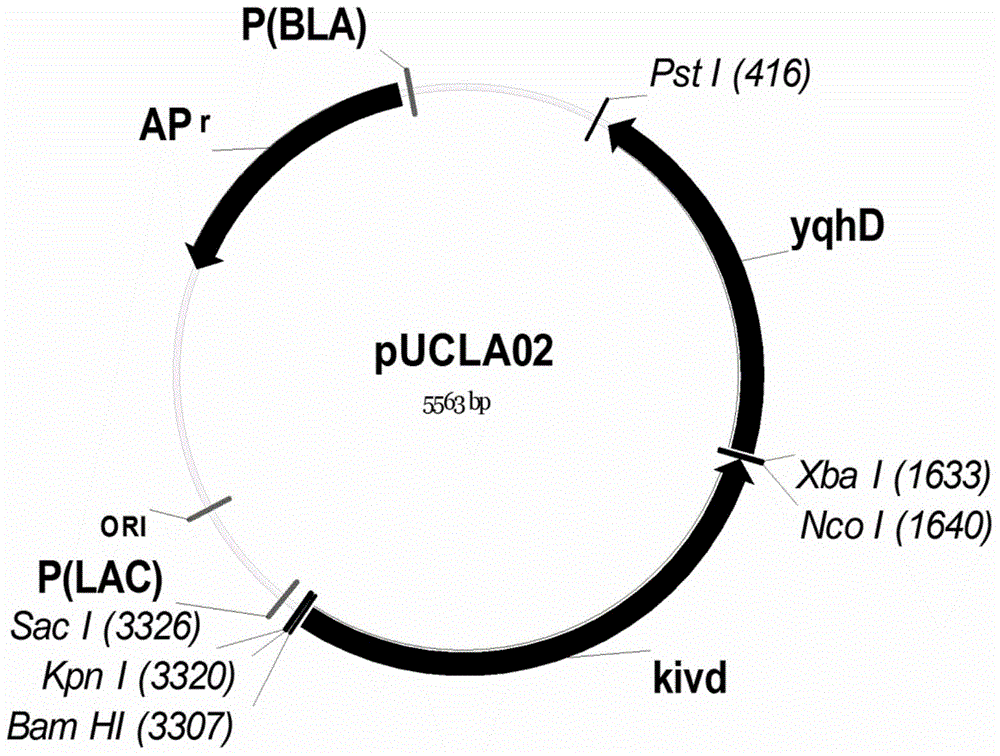
[0068] The recombinant plasmid pACYCLA09 and pTRCLA10 construction of embodiment 2 isobutanol synthesis pathway
[0069] (1) First extract the DNA template
[0070] Bacillus subtilis 168 was cultured in LB medium (10g / L peptone, 5g / L yeast powder, 10g / L NaCl, pH 7.0) at 37°C and 200rpm, and after reaching the logarithmic phase, use the genome extraction kit (Tiangen) Extract total DNA.
[0071] Lactococus lactis was cultured in MRS medium (10g / L peptone, 10g / L beef extract powder, 5g / L yeast powder, 20g / L glucose, 1.0mL Tween 80, 2g / L diammonium hydrogen citrate, 5g / L sodium acetate , 2g / L K 2 HPO 4 ·3H 2 O, 0.58g / L MgSO 4 ·7H 2 O, 0.25g / LMnSO 4 ·H 2 0, pH 7.0) to the logarithmic phase after static culture, the genome extraction kit (Tiangen) was used to extract the total DNA.
[0072] E.coli MG1655 was cultured in LB medium (10g / L peptone, 5g / L yeast powder, 10g / L NaCl, pH 7.0) at 37°C and 200rpm, and after reaching the logarithmic phase, the genome extraction kit (T...
Embodiment 3
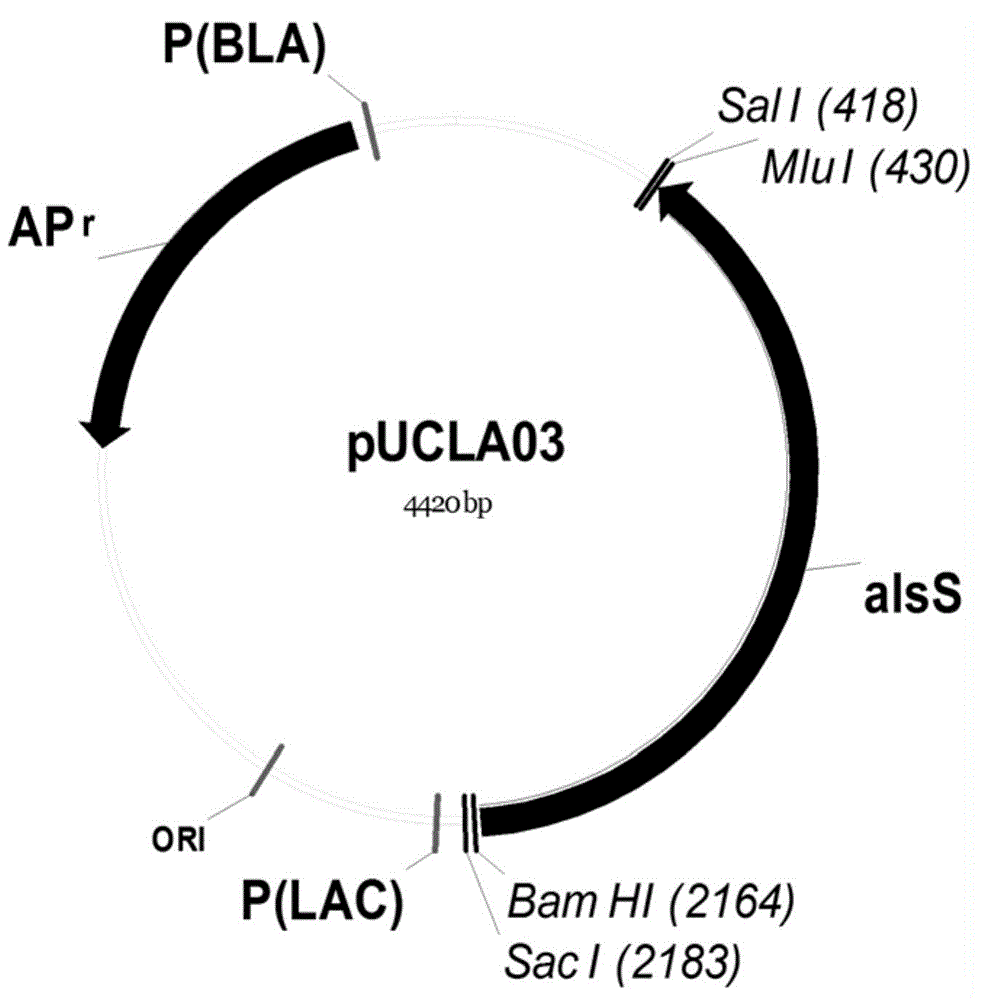
[0088] Embodiment 3 initial isobutanol synthesis bacterial strain is constructed
[0089] (1) Escherichia coli E.coli LA01 electroporation competent preparation
[0090] 1) Inoculate the strains transferred by streaking in -80 refrigerator, and culture at 37°C.
[0091] 2) Pick a single clone, transfer it to a 5 mL LB test tube, culture at 200 rpm, 37° C. overnight.
[0092] 3) Inoculate 1% inoculum into 50ml LB medium, 37°C, 200rmp, OD 600 Stop culturing when it reaches 0.5-0.6.
[0093] 4) Under sterile conditions, transfer the bacteria to an ice-precooled 50mL centrifuge tube, place on ice for 20min, centrifuge at 4200rpm, 4°C for 10min.
[0094] 5) Discard the medium, invert the tube for 1 min, resuspend the pellet with 50 mL pre-cooled 10% glycerol, and centrifuge and wash 3 times.
[0095] 6) Add 500uL of 10% glycerol to resuspend, and distribute in sterilized precooled centrifuge tubes, about 100uL in each centrifuge tube.
[0096] 7) Rapidly cooled with liquid nit...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com