Escherichia coli genome integration vector, genetically engineered bacterium and application of genetically engineered bacterium to xylitol production
A technology of genetically engineered bacteria and integrated vectors, applied in the fields of genetic engineering and biology, can solve the problems of time-consuming and laborious, cumbersome experimental operation, and reduced recombination efficiency, and achieve the effect of solving time-consuming and laborious, simplifying integration technology, and reducing production costs.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Example 1 Construction of genetically engineered bacteria with different promoters
[0045] 1. Construction of Trc promoter expression vector
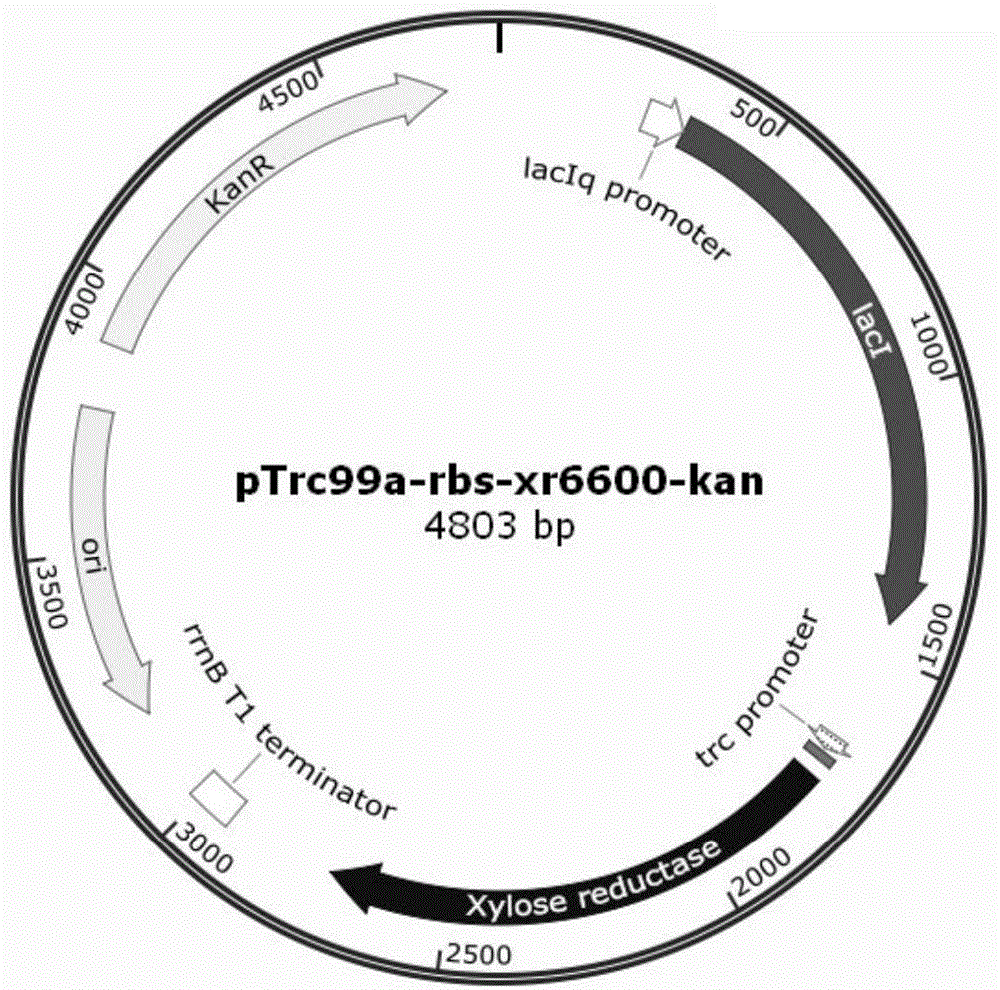
[0046] Using plasmids pET-30a(+) and pTrc99a-rbs-xr6600 as templates, primers ori-kan-P1 and ori-kan-P2 were designed to amplify the replicon and kanamycin resistance gene on pET-30a(+) , design primers Ptac+XR-P1 and Ptac+XR-P2 to amplify the promoter, target gene and terminator on the plasmid pTrc99a-rbs-xr6600, digest with enzymes respectively, connect with T4 ligase after digestion, and construct a recombinant plasmid; Transform the recombinant plasmid into Escherichia coli competent cell DH5α, pick the colony grown on the kanamycin plate, extract the plasmid and sequence it, and name the correct recombinant plasmid as follows:
[0047] pTrc99a-kan-xr6600, the plasmid was transferred into strain HK401, and a genetically engineered bacterium containing the corresponding recombinant plasmid was constructed, which was named HK...
Embodiment 2
[0078] Example 2 Test of different engineering bacteria producing xylitol ability
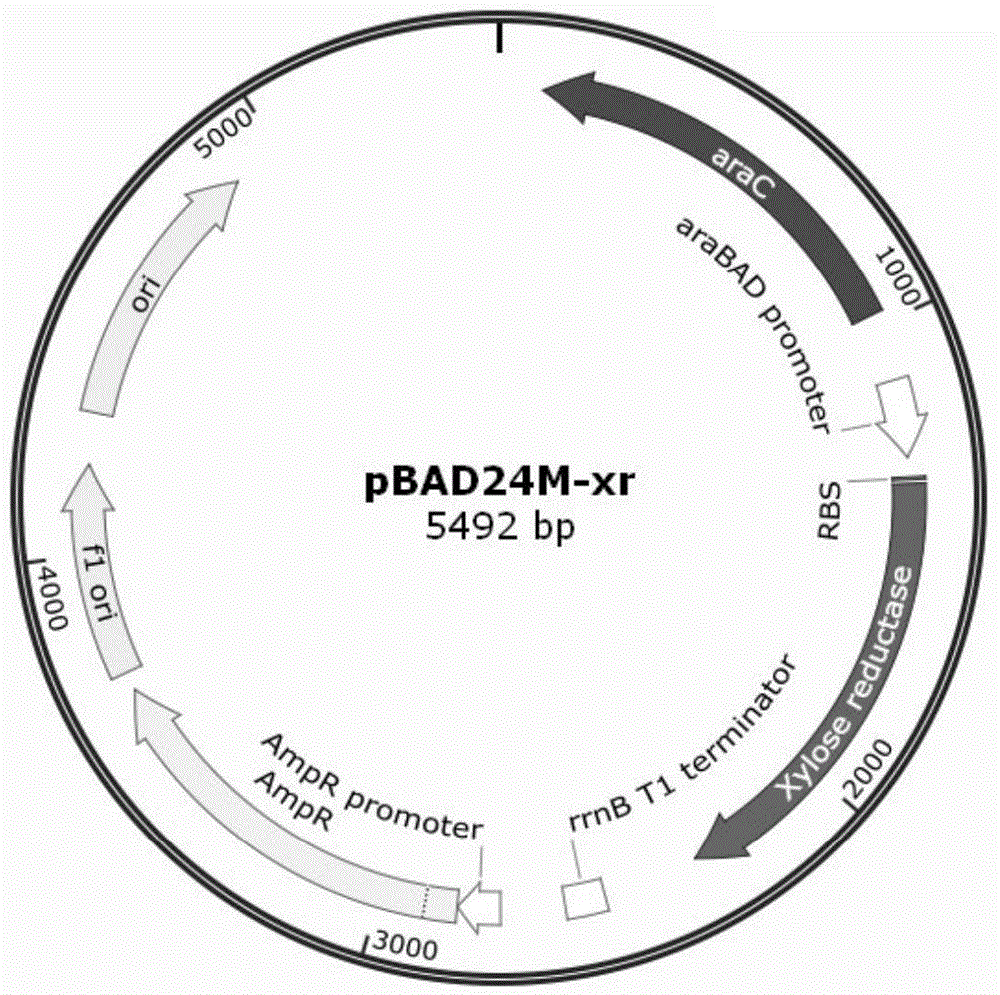
[0079] Inoculate engineering bacteria HK412, HK422 and HK432 in 45mL of improved M9 medium (1L medium contains 4-6g Na 2 HPO 4 , 2~5g KH 2 PO 4 , 1~2g NH 4 Cl, 1~5g NaCl, 1~5mM MgSO 4 , 1~5mM CaCl 2 , 2~10g / L yeast extract), cultured at 30°C until OD600 was 0.6~1, added an appropriate amount of inducer (Trc: IPTG pBAD: arabinose) and added xylose to the fermentation broth to a final concentration of 20g / L, adding glucose to a final concentration of 10g / L, culturing at 30°C, investigating the fermentation characteristics of each engineered bacteria, the results of the investigation are as follows Figure 5 , 6 , 7 shown.
[0080] Such as Figure 5 As shown, the engineered bacteria HK412 can consume glucose and xylose within 34.5 hours after being induced by 0.1mM IPTG, produce xylitol 18.5g / L, and the production efficiency is 0.62g / L / h;
[0081] Such as Image 6 As shown, under the i...
Embodiment 3
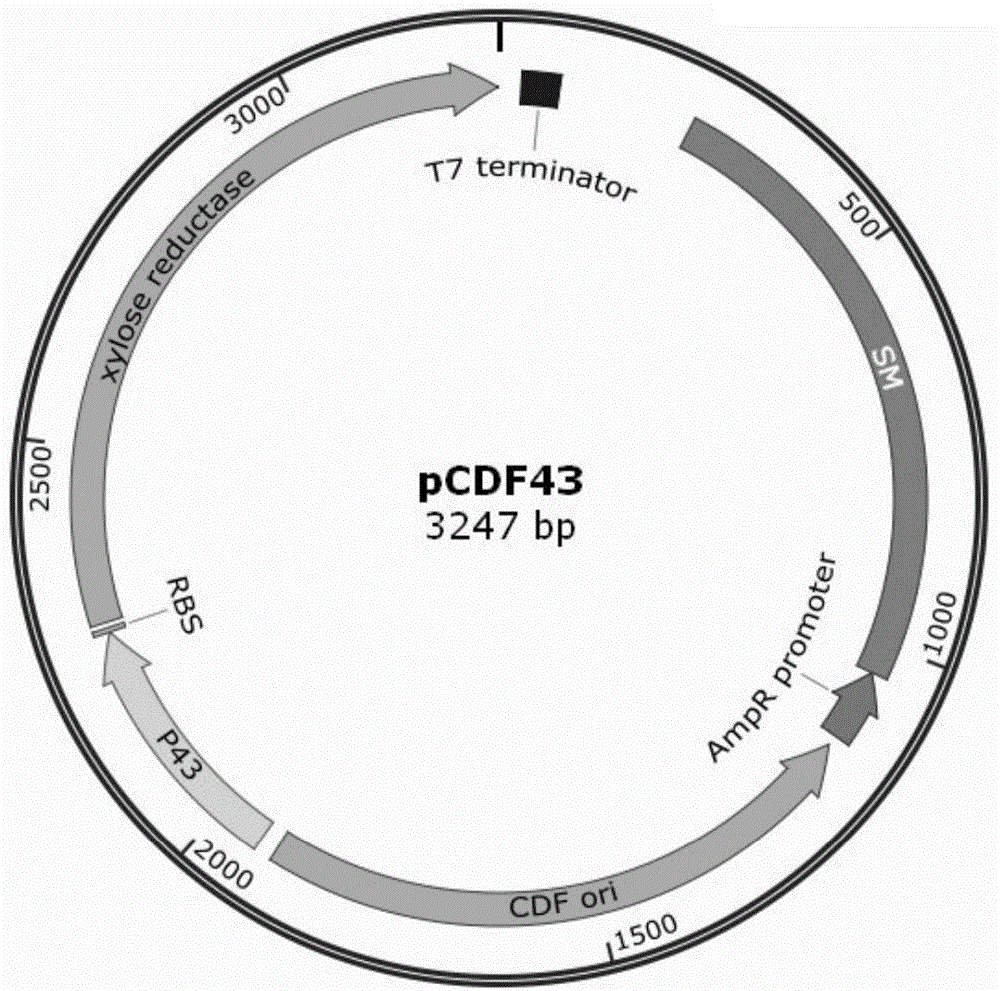
[0083] Example 3 Construction of integration vector
[0084] Use primers CM+R6K-P1 and CM+R6K-P2 to amplify the R6K replicon and the chloramphenicol resistance gene carrying the FRT site using PKD3 as a template, and use primers IS5-P1 and IS5-P2 to amplify the insert sequence IS5, Link the IS5 sequence with the R6K replicon and chloramphenicol resistance by overlapping PCR, design primers pCDF43-P1 and pCDF43-P2, use pCDF43 as a template to amplify the promoter P43, XR and terminator, respectively digest, enzyme After cutting, connect with T4 ligase to construct a recombinant plasmid; transform the recombinant plasmid into Escherichia coli competent cell DH5α, pick the colonies grown on the streptomycin sulfate plate, extract the plasmid and sequence it, and verify the correct naming of the recombinant plasmid For: pRC43.
[0085] Wherein, the concrete sequence of primer is as follows:
[0086] CM+R6K-P1: 5'-AAAA CTGCAG AGTAGGGAACTGCCAGGCATCAA-3'
[0087] CM+R6K-P2: 5’-A...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com