Group of genes for lung cancer molecular subtyping and application thereof
A molecular typing and genetic technology, applied in the field of cancer diagnosis and molecular biology, can solve the problems of high false positive and false negative, low diagnostic efficiency, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0073] Training set sample collection and processing:
[0074] The present invention analyzes the clinical data and biological sample data of lung cancer patients with a large sample size, including 391 cases of lung adenocarcinoma and 237 cases of lung squamous cell carcinoma, and a total of 628 cases of relevant clinical data and gene expression data of patients to construct a lung cancer gene expression profile database.
[0075] Screening of 13 specific genes:
[0076] According to the measured value of gene expression abundance, the inventors used the statistical analysis method T-test to screen out 13 genes that are closely related to the primary tumor site from more than 20,000 genes. These genes were differentially expressed in lung cancer subtypes, with statistical significance, as shown in Table 1.
[0077] Table 1: 13 Gene Sets
[0078] Gene
note
P value
COMP
Cartilage Oligomeric Matrix Protein
<0.001
FXYD3
FXYD Doma...
Embodiment 2
[0082] Validation set test:
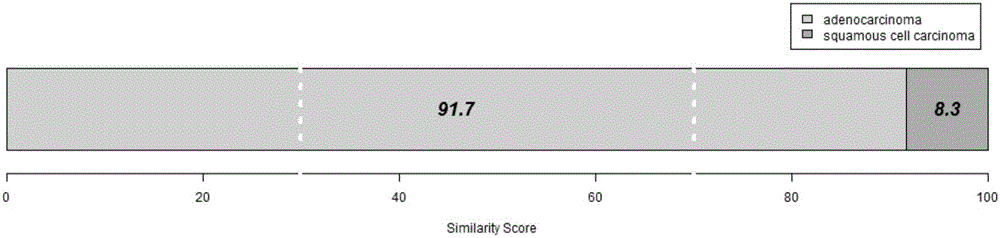
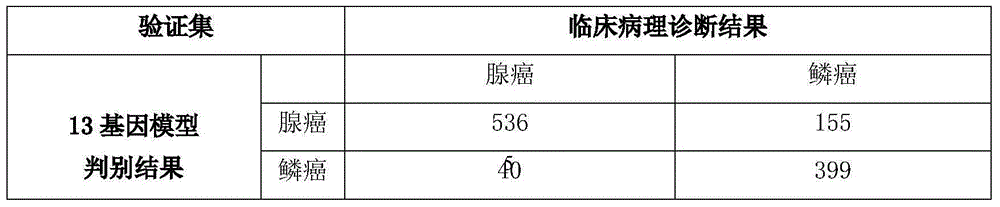
[0083] In this example, the inventors analyzed the high-throughput sequencing data of 1130 cases of lung cancer, including 576 cases of lung adenocarcinoma and 554 cases of lung squamous cell carcinoma. The 13-gene statistical analysis model was used to discriminate the subtype of each sample, and compared with the results of clinicopathological diagnosis, the accuracy rate was 82.7%. Taking lung adenocarcinoma as a reference, the predicted sensitivity is 93.0%, and the specificity is 72.0%, see Table 2.
[0084] Table 2: Discrimination results of the 13-gene model in the validation set of 1130 cases
[0085]
Embodiment 3
[0087] Screening of 30 specific genes:
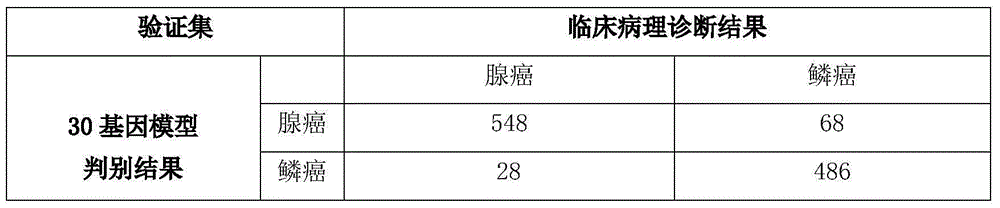
[0088] The inventors relaxed the screening criteria of the T-test to a P value of less than 0.01, and further obtained 17 additional genes. These genes were differentially expressed in lung adenocarcinoma and lung squamous cell carcinoma, and the difference was statistically significant. The 17 genes were combined with the 13 genes in Example 1 to form a set of 30 genes, as shown in Table 3.
[0089] Table 3: 17 Gene Sets
[0090] Gene
note
P value
KRT5
Keratin 5, Type II
<0.01
S100A2
S100A2
<0.01
KRT6A
Keratin 6A, Type II
<0.01
MIR205HG
MIR205 Host Gene
<0.01
KRT17
Keratin 17, Type I
<0.01
CLCA2
Chloride Channel Accessory 2
<0.01
SERPIN B3
Serpin Peptidase Inhibitor, Clade B (Ovalbumin), Member 3
<0.01
MMP12
Matrix Metallopeptidase 12
<0.01
DSG3
Desmoglein 3
<0.01
KRT6B
Keratin 6B...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com