High-flux molecular marker for identifying pulp color of Cucumis melo L. and special primer for high-flux molecular marker
A technology for melon and pulp, applied in the field of molecular biology, can solve problems such as cumbersome processes, achieve high analytical throughput, reduce human errors, and speed up transformation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Example 1. High-throughput KASP marker design of the gene CmOr related to the color and shape of melon pulp and development of its special primer sequence
[0043] 1. Extraction of tested muskmelon materials and their genomic DNA
[0044] A. Selection of test materials
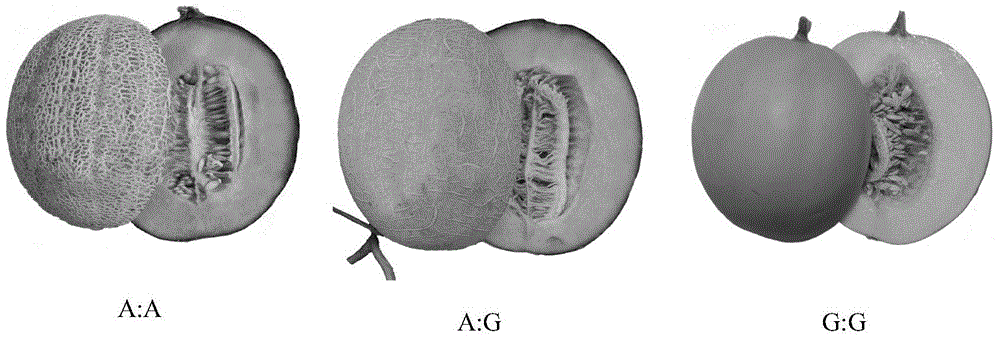
[0045] Test material: orange-red flesh melon M285 (A:A genotype), green-flesh melon Lubao (G:G genotype), and white-flesh melon Elizabeth (G:G genotype) with known genotypes of the CmOr gene. Among them, the genotypes of the CmOr genes of these three melons have been sequenced and verified.
[0046] B. Genomic DNA extraction
[0047] The test materials were subjected to DNA extraction treatment respectively to obtain the genomic DNA of the test materials;
[0048] DNA extraction was improved on the basis of the method (MurrayM, ThompsonWF.RapidisolationofhighmolecularweightplantDNA[J].NuclAcidRes,1980,8:668-673.) with reference to Murray et al. (1980); the specific steps are as follows:
[0049] (1)...
Embodiment 2
[0078] Example 2, the establishment of a method for detecting the CmOr gene of muskmelon using high-throughput KASP markers
[0079] 1. Genomic DNA extraction
[0080] See Step 1 of Example 1.
[0081] 2. PCR amplification
[0082] Using the genomic DNA extracted in step 1 as a template, the special primers developed in Example 1 for detecting the KASP marker of the melon CmOr gene were used for PCR amplification respectively to obtain PCR amplification products.
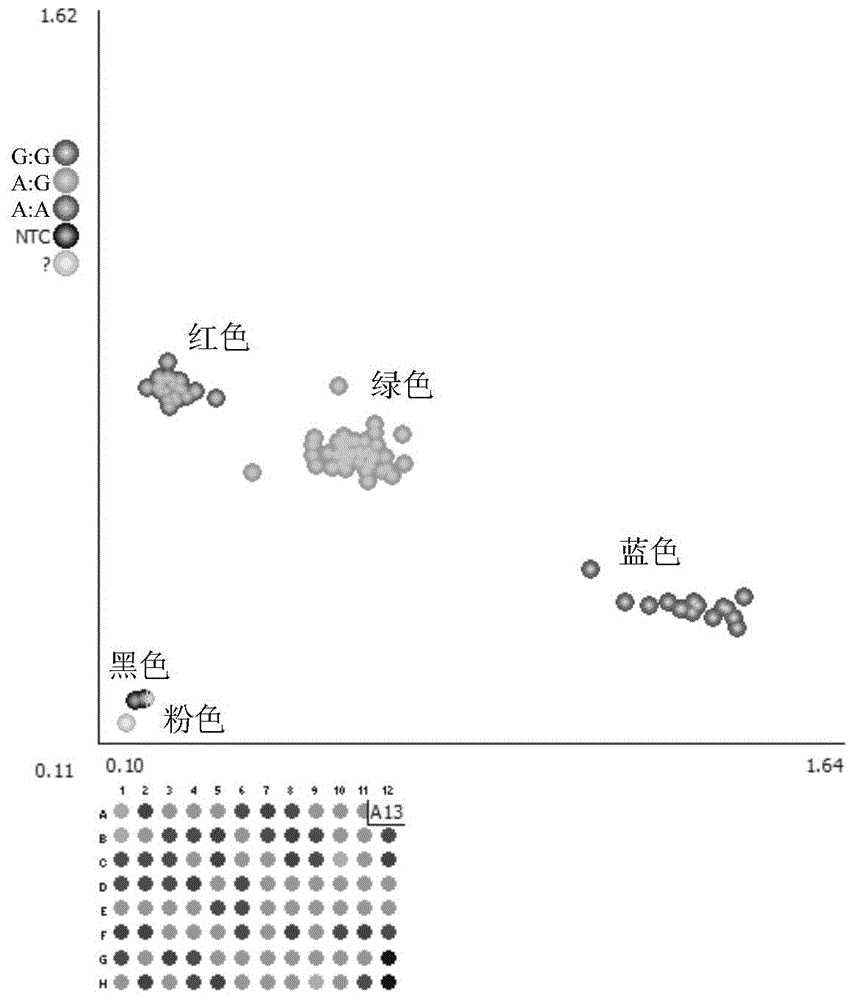
[0083] KASP genotyping PCR reaction system:
[0084] 96-well plate: 10ng genomic DNA, 5μl KASPV4.02×MasterMix, 0.14μl KASP72×assaymix, add ddH 2 0 to 10 μl.
[0085] 384-well plate: 5ngDNA, 2.5μl KASPV4.02×MasterMix, 0.07μl KASP72×assaymix, add ddH 2 0 to 5 μl.
[0086] 1536-well plate: 5ngDNA, 2.5μl KASPV4.02×MasterMix, 0.07μl KASP72×assaymix, add ddH 2 0 to 5 μl.
[0087] Among them, KASPV4.02×MasterMix is a product of LGC Company, and the catalog number of KASPV4.02×MasterMix for 96 / 384-well plates is K...
Embodiment 3
[0098] Example 3. Verification of the high-throughput KASP marker detection of the CmOr gene in melon and its application in breeding
[0099] 1. Test materials
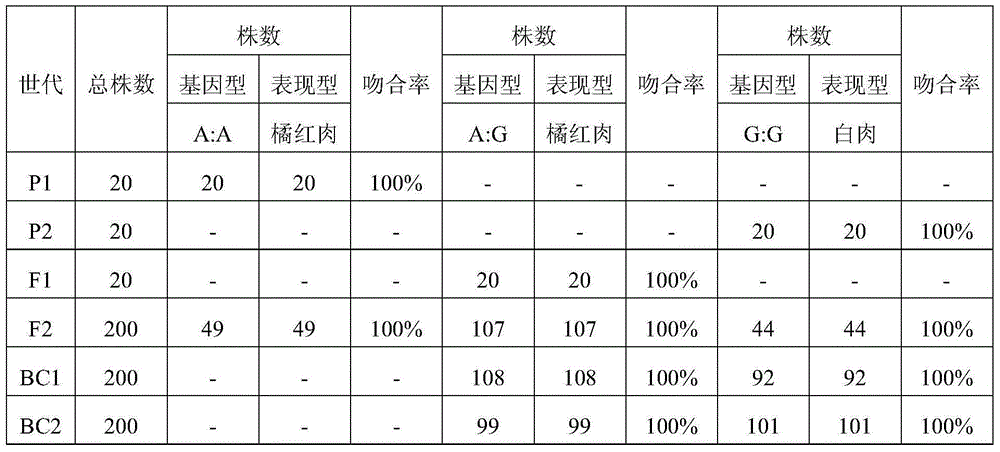
[0100] The test materials include: the orange-red flesh melon variety M285 (A:A genotype) is used as the donor parent, and the white-flesh melon variety Elizabeth (G:G genotype) is used as the recurrent parent, and the color traits of the melon flesh are backcrossed. M285 was crossed with Elizabeth to obtain the corresponding F1 generation, the F1 generation was self-crossed to obtain the F2 population, and the F1 was backcrossed with the recurrent parent Elizabeth to obtain the BC population. The orange-red flesh melon varieties PI414723 (A: A genotype) and Védrantais (A: A genotype), the green flesh melon variety Lubao (G: G genotype), the parents Elizabeth (G: G genotype) and M285 (A : A genotype) as a control for molecular marker-assisted marker screening. The genotype of the CmOr gene of the control melon has ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com