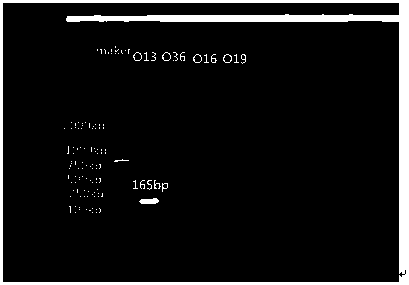
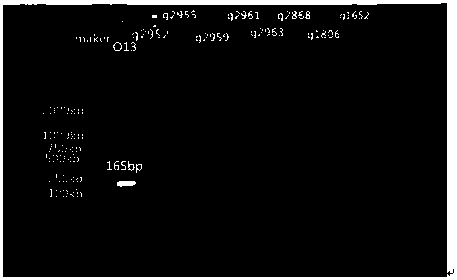
Nucleotides specific to Aeromonas hydrophila o13, o36, o16 and o19 and their application
A hydrophilic Aeromonas, nucleotide technology, applied to the hydrophilic Aeromonas O13, can solve the problems of high missed detection rate, low sensitivity, insufficient quantity, etc., to achieve high accuracy, low detection cost, Practical effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Example 1 : Genome Extraction
[0037] Aeromonas hydrophila was cultivated in nutrient broth medium at 37°C, the bacteria were collected, and the genome was extracted. The specific steps are as follows:
[0038] The cells were resuspended with 500ul 50mM Tris-HCl (pH8.0) and 10ul 0.4M EDTA, incubated at 37°C for 20 minutes, and then 10ul 10mg / ml lysozyme was added to continue the incubation for 20 minutes. Then add 3ul 20mg / ml proteinase K, 15ul10% SDS, incubate at 50°C for 2 hours, then add 3ul 10mg / ml RNase, and incubate at 65°C for 30 minutes. Add an equal volume of phenol to extract the mixture, take the supernatant, and then extract twice with an equal volume of phenol: chloroform: isoamyl alcohol (25:24:1), take the supernatant, and then extract with an equal volume of ether. Extract to remove residual phenol. Precipitate DNA with 2 times the volume of ethanol in the supernatant, roll out the DNA with glass wool and wash the DNA with 70% ethanol, and finally r...
Embodiment 2
[0039] Example 2: sequence deciphering
[0040] Extract the genomes of the standard strains of each serotype of Aeromonas hydrophila, and use Solexa pair-end sequencing technology to perform whole-genome sequencing on the genomes of each serotype of Aeromonas hydrophila to obtain the sequence of the serotype, using Blast and PSI-Blast Sequence alignment was carried out, TMHMM 2.0 program was used for transmembrane structure prediction, and ClustalW program was used for sequence alignment and screening of conserved and specific gene fragments, and finally the O antigen gene cluster sequence and deciphering results of each serotype of Aeromonas hydrophila were obtained.
Embodiment 3
[0041] Example 3 : Primer design
[0042] The O antigen gene cluster sequences of each serotype of Aeromonas hydrophila were self-tested by our laboratory. Through comparative analysis, we selected gene-specific segments with relatively low identity and similarity values in the Blast comparison results to design primers. Of which O13 serotype wxya The identity value and similarity value of gene comparison result are 57% and 76%; the identity value and similarity value of wzm gene comparison result of O36 serotype are 73% and 83%; the identity value and similarity value of wzx gene comparison result of O16 serotype Values are 61% and 78%; the identity value and similarity value of the GT gene comparison results of O19 serotype are 84% and 91%; so each serotype selects the above-mentioned corresponding gene as the specific target gene of the serotype, for each serotype Specific primers were designed for the specific segments of the genes of the serotypes.
[0043] Prime...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com