Fluorescent quantitative PCR method for detecting fish parvalbumin and primer pair
A technology of parvalbumin and fluorescence quantification, applied in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc. problems, to achieve the effect of low detection cost, simple result judgment, and reliable detection results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
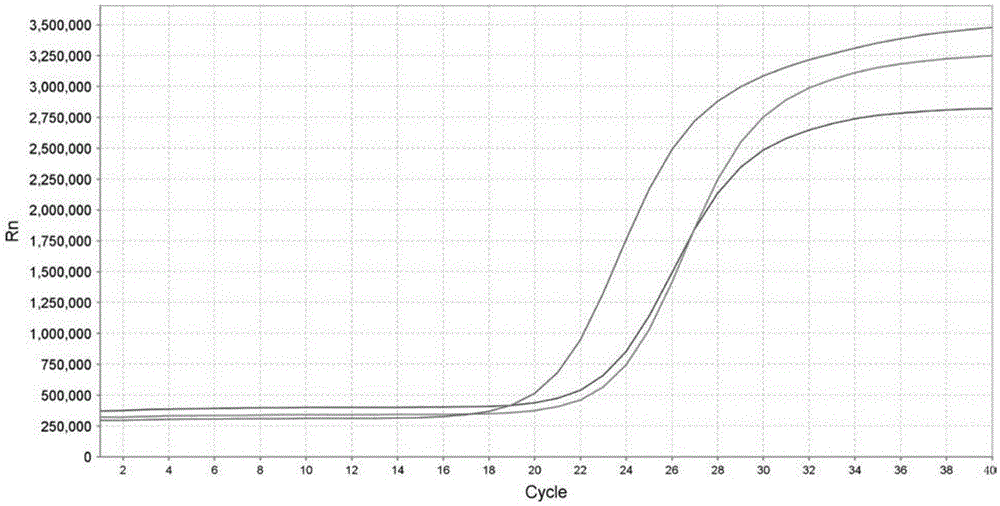
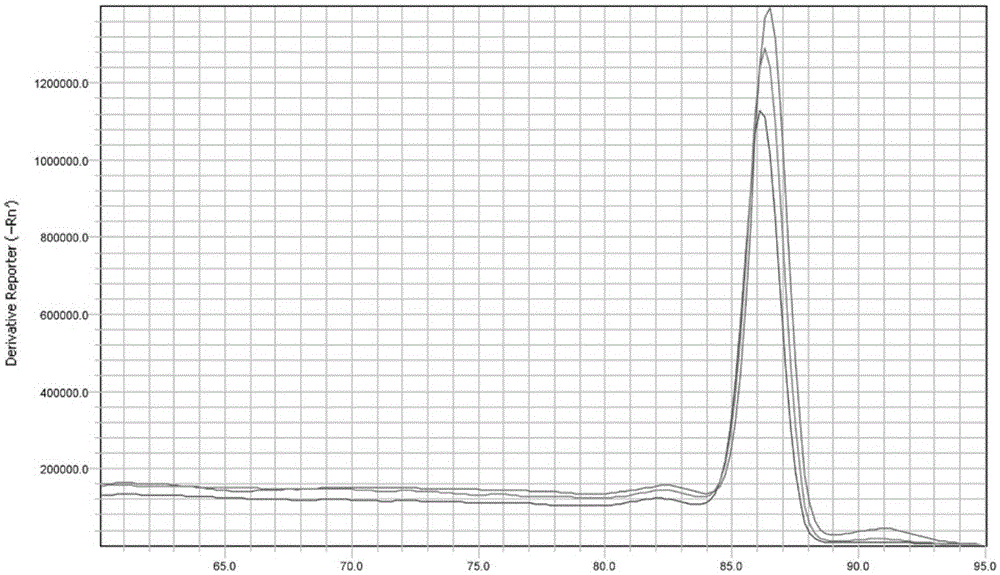
[0023] Embodiment 1: detect the real-time fluorescent quantitative PCR method of perch parvalbumin, specifically comprise the following steps:
[0024] Step 1, designing primers according to the conserved sequence in the genome DNA sequence encoding fish parvalbumin
[0025] Through the analysis of amino acid sequence and coding gene sequence, because fish parvalbumin has good conservation, primers were designed according to the specificity of the protein coding gene. First find the parvalbumin-encoding genes of different fish in GenBank, and find out the conserved segments after comparison analysis, and then compare the selected conserved segments with the DNA sequences of other animal species in GenBank by Blast software, Find the non-homologous region of DNA with other species, and use the primer design software PrimerPremier5.0 to design specific primers for parvalbumin therefrom. The primer sequence is as follows (primers are provided by Dalian Bao Biological Engineering ...
Embodiment 2
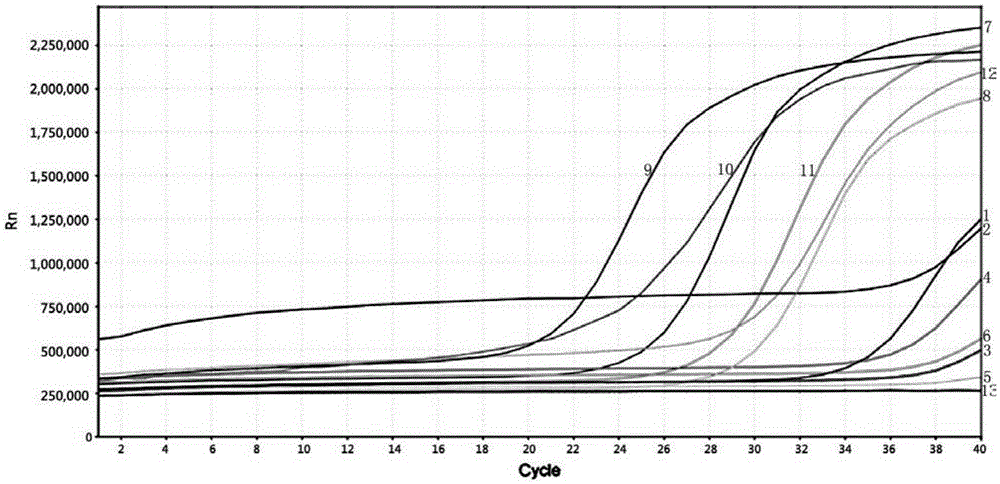
[0038] Embodiment 2: Utilize the detection method of embodiment 1, to chicken, beef, donkey meat, mutton, duck, pork 6 kinds of non-fish meat products, and small yellow croaker, large yellow croaker, salmon, sea bream, red fish fillet, cod 6 kinds of fish products were tested for parvalbumin gene, the results are as follows image 3 , 4As shown (1-12 in the figure respectively represent: 1. Chicken; 2. Beef; 3. Donkey meat; 4. Mutton; 5. Duck; 6. Pork; 7. Small yellow croaker; 8. Large yellow croaker; 9. Salmon ; 10. sea bream; 11. red fish fillet; 12. cod), 6 kinds of non-fish meat products were all negative; the test results of the remaining 6 kinds of fish meat products were all positive.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com