Method for detecting pseudomonas fluorescens and kit and primers thereof
A technology of Pseudomonas fluorescens and a kit, which is applied in the field of microbial detection, can solve the problems of high false positives and false negatives, long detection time, complicated operation, etc. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
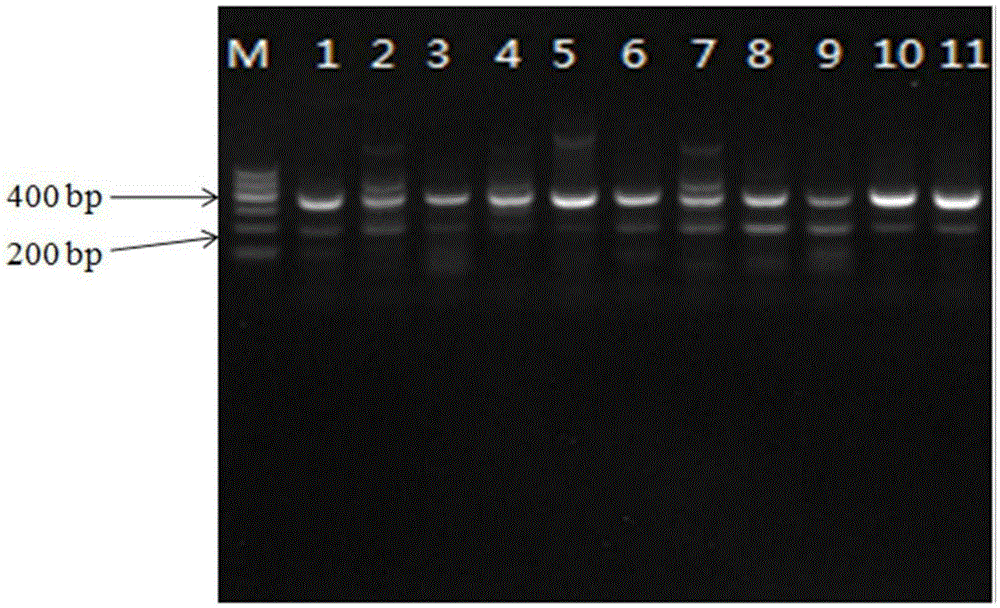
[0030] Embodiment 1 is detected to the double PCR of Pseudomonas fluorescens bacterial strain
[0031] (1) Primer synthesis
[0032] Synthesize primers capable of PCR amplifying Pseudomonas fluorescens gyrase (gyrase) subunit gene and metalloproteinase operon (apr) specific gene sequence, the sequence is as follows:
[0033] SEN1-L: 5'-GCCAGCAGAGTCACCTTCCA-3' (SEQ ID NO: 1),
[0034] SEN2-R: 5'-AGCACAAAGTCGCCACCACC-3' (SEQ ID NO: 2),
[0035] SEN3-L: 5'-TAYGGBTTCAAYTCCAAYAC-3' (SEQ ID NO: 3),
[0036] SEN4-R: 5'-VGCGATSGAMACRTTRCC-3' (SEQ ID NO: 4).
[0037] SEN3-L and SEN4-R are degenerate primers, Y, B, V, M, S and R are all degenerate bases, where Y represents C or T, B represents G, C or T, V represents A, G Or C, M for A or C, S for G or C, R for A or G.
[0038] (2) Reaction system and reaction parameters of double PCR detection method
[0039] Using the above primers, using the genomic DNA of Pseudomonas fluorescens strain as a template, the PCR reaction system an...
Embodiment 2
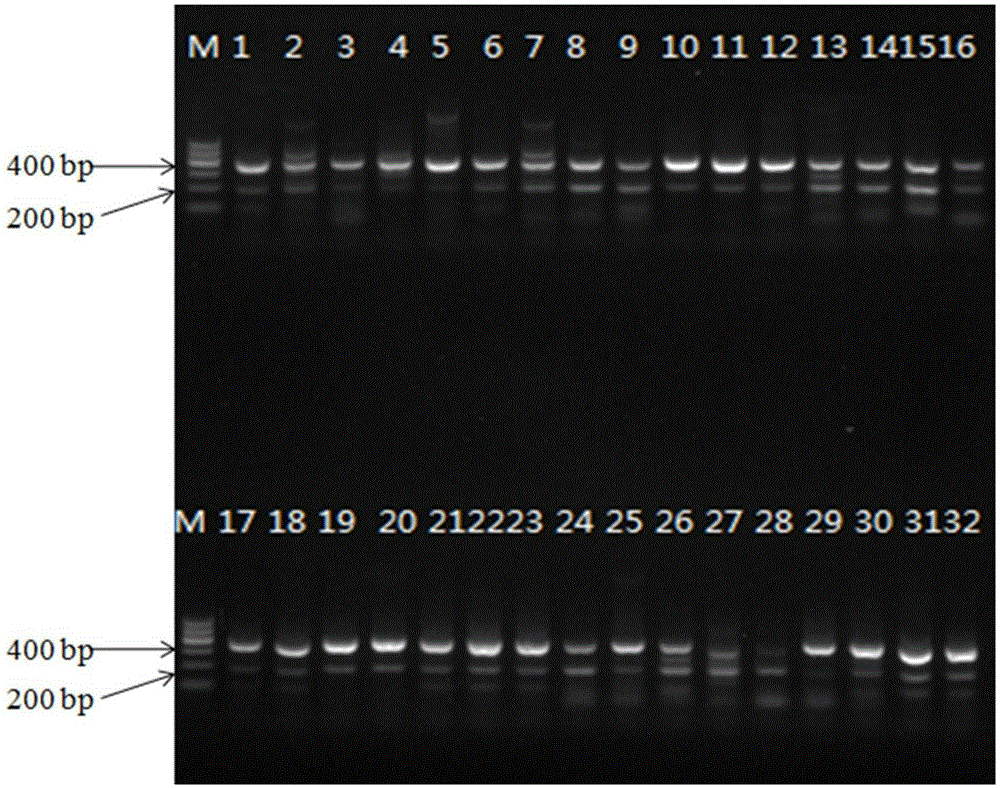
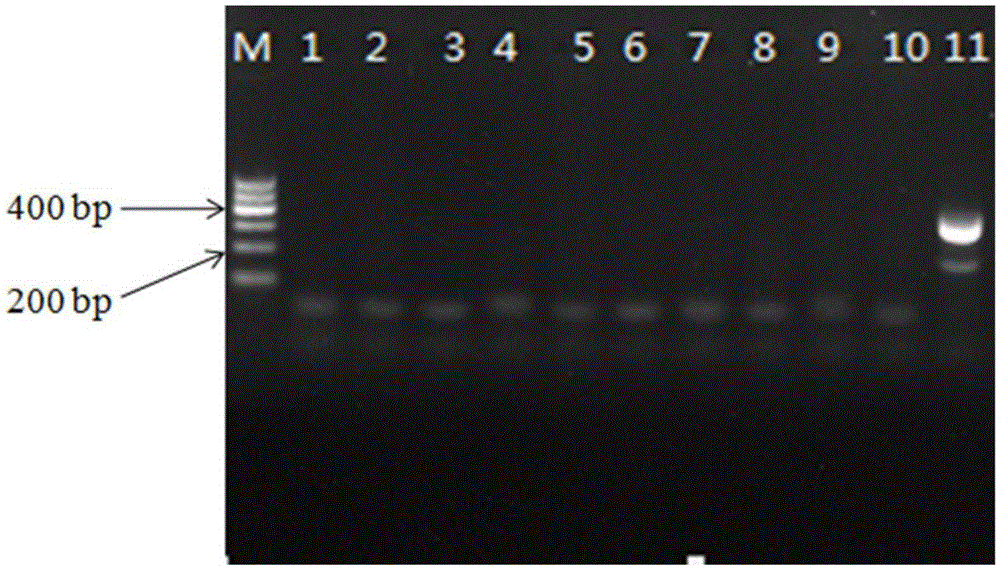
[0062] Example 2 Detection test of artificial contamination Pseudomonas fluorescens model strain AS1.867
[0063] The activated Pseudomonas fluorescens (AS1.867) bacteria solution was inserted into LB medium and cultured on a shaker until the stationary phase, and the initial bacterial concentration was 1.8×10 9 cfu / mL. Add 25mL of LUHT milk samples, that is, milk that has undergone ultra-high temperature instant sterilization (135°C, 3-5s), into 225mL of selective enrichment solution (selective enrichment solution includes the following components: peptone 10g, tryptone 10g, yeast extract 10g, NaCl 10g, distilled water to 1L), respectively insert 1mL of Pseudomonas fluorescens with concentrations of 180cfu / mL, 18cfu / mL, and 1.8cfu / mL, and do 3 parallel experiments for each concentration. Samples were taken once at 0h, 4h, 6h, 8h, 10h, 12h, and 24h, and genomic DNA was extracted by boiling method. 2 μL of samples at each time point was added as a PCR reaction template to the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com