Recombinant strain for producing glycolate based polymers and application of recombinant strain
A recombinant bacteria and biosynthesis technology, applied in the biological field, can solve the problems of inability to study material properties, low biomass, difficult polymer extraction, etc., and achieve the effect of good industrial application prospects.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
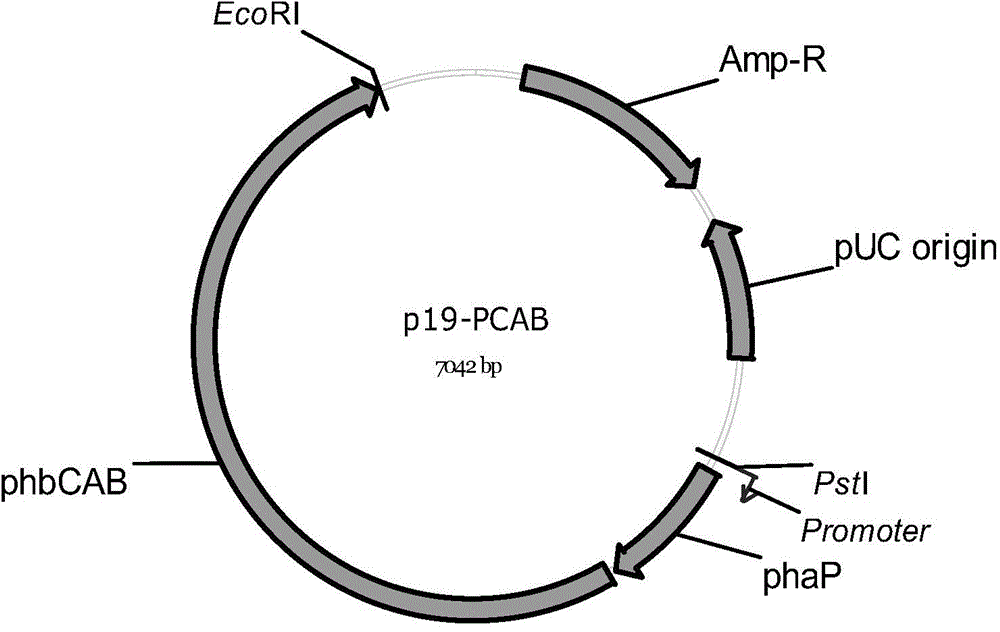
[0071] Embodiment 1, the construction of recombinant expression vector p19-PCAB
[0072] 1. Artificially synthesize the DNA shown in sequence 1 in the sequence list, wherein the 13th-55th nucleotides are the promoter sequence, the 72nd-650th nucleotides are the phaP gene sequence, and the 682-4395th nucleotides are phbCAB operator sequence.
[0073] 2. Digest the DNA sequence synthesized in step 1 with PstI and EcoRI, and recover a gene fragment with a size of about 4.4 kb.
[0074] 3. The vector pMD19-T (purchased from Takara, Japan) was double-digested with PstI and EcoRI, and a vector fragment with a size of about 2.6 kb was recovered.
[0075]4. The gene fragment obtained in step 2 is connected with the carrier fragment obtained in step 3 (solutionI connection kit of Takara), and the connection product is imported into E.coliJM109 (Promega Biotechnology Co., Ltd., catalogue) by chemical transformation No. P9751), spread on LB-Amp solid medium, and culture at 37°C for 16 ...
Embodiment 2
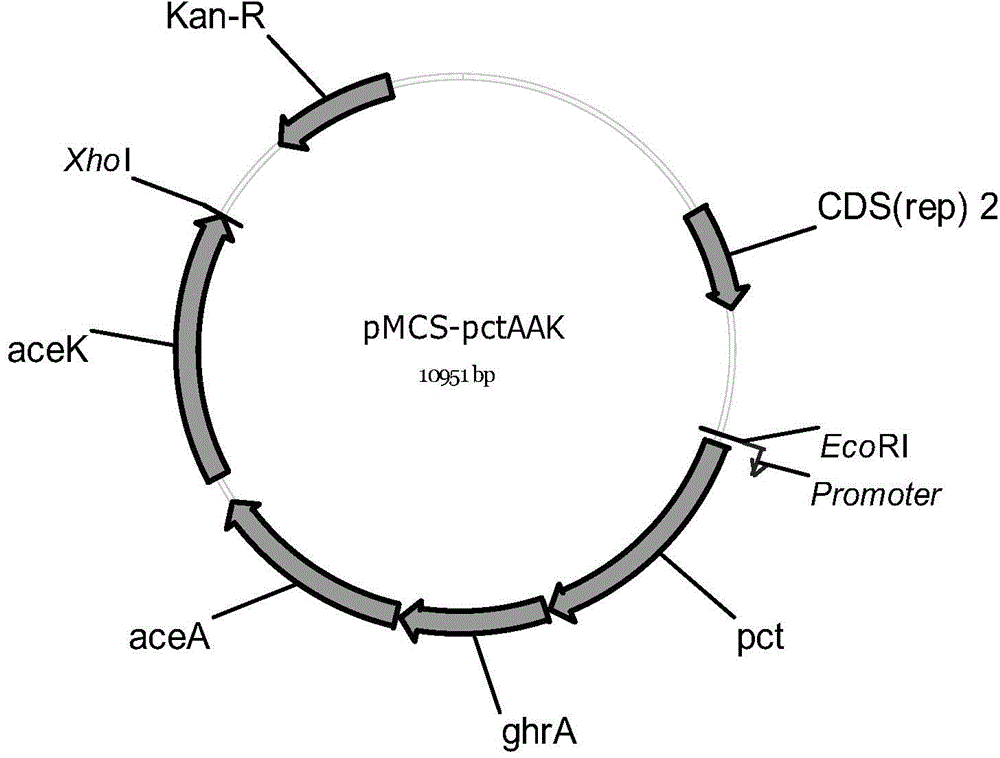
[0078] Embodiment 2, construction of recombinant expression vector pMCS-pctAAK
[0079] 1. The artificially synthesized DNA sequence shown in Sequence 2 in the sequence listing, wherein the 13th-55th nucleotides are the promoter sequence; the 72nd-1625th nucleotides are the pct gene sequence; the 1655th-2593rd nucleotides It is the ghrA gene sequence; the 2623-3927th nucleotide is the aceA gene sequence; the 4110-5826th nucleotide is the aceK gene sequence;
[0080] 2. Digest the DNA sequence synthesized in step 1 with EcoRI and XhoI, and recover a gene fragment with a size of about 5.8 kb;
[0081] 3. Digest the vector pBBR1MCS-2 with EcoRI and XhoI (the public can synthesize it by themselves according to the sequence of NCBI GenBank No. U23751), and recover the vector fragment with a size of about 5.1kb;
[0082] 4. Ligate the gene fragment obtained in step 2 with the carrier fragment obtained in step 3 (Takara's solutionI connection kit), and the obtained connection produc...
Embodiment 3
[0085] Embodiment 3, the construction of escherichia coli JM109ldhA
[0086] 1、合成ldhA基因敲除所用的引物ldhAF:5'-CTCCCCTGGAATGCAGGGGAGCGGCAAGATTAAACCAGTTCGTTCGGGCAATTCCGGGGATCCGTCGACC-3'和ldhAR:5'-TATTTTTAGTAGCTTAAATGTGATTCAACATCACTGGAGAAAGTCTTATGTGTAGGCTGGAGCTGCTTCG-3',以质粒pKD13为模板,采用ldhAF和ldhAR引物PCR扩增得到1.5kb左右的DNA Fragment, the sequence of the fragment is shown in SEQIDNo.3, and the obtained DNA fragment is purified by agarose gel electrophoresis;
[0087] 2. Cultivate Escherichia coli containing pKD46 in LB-Amp liquid medium at 30°C, extract plasmid pKD46, transform it into recipient strain E.coliJM109 by electroporation, and spread it on LB-Amp solid culture medium at 30°C for 24 hours to obtain a transformant, E.coliJM109(pKD46);
[0088] 3. Inoculate E.coliJM109(pKD46) into LB-Amp liquid medium, incubate at 30°C for 4h, add arabinose to a final concentration of 20g / L, continue to incubate for 1.5h, and then prepare the E.coliJM109(pKD46) sensation For competent cells, the DNA frag...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com