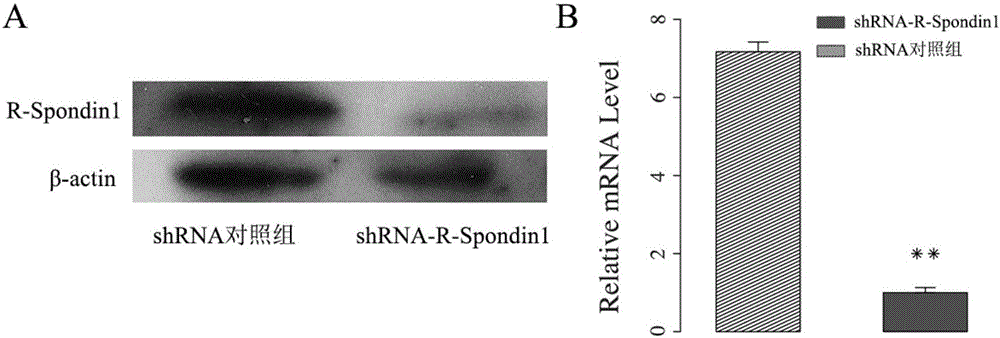
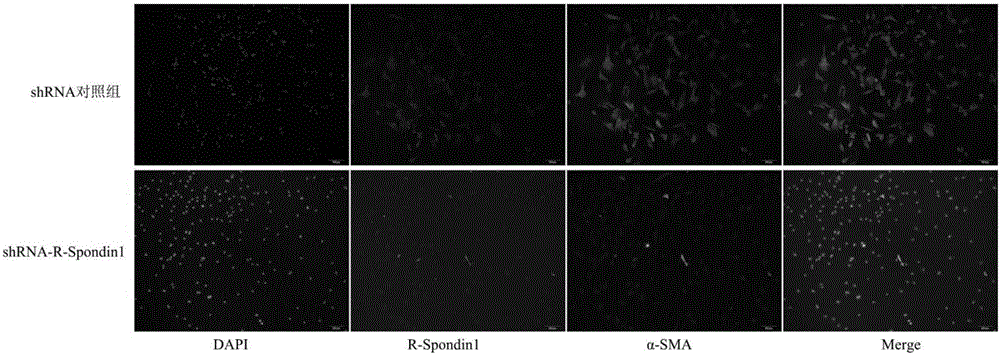
Small interfering RNAs, short hairpin RNAs, vectors and applications targeting mammalian r-spondin1 gene targets
A mammalian, small-interference technology, applied in the direction of DNA/RNA fragments, vectors, nucleic acid vectors, etc., can solve the problems of short-term inhibition of target gene expression, non-persistent expression of siRNA, low transfection efficiency, etc., to promote the recovery process. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Example 1 siRNA sequence design
[0032] Apply the principles of siRNA design (Petri S, siRNA design principles and off-target effects. Methods Mol Biol. 2013; 986:59-71) to screen siRNA sequences: (1) start from the AUG start codon of the transcript (mRNA), search for "AA" double-linked sequence, and write down the 19-23 nucleotide sequence at its 3' end as the candidate target site of siRNA; (2) the GC content of the siRNA sequence is between 45% and 55%; (3) the length 19-23 bp; (4) no inverted repeat sequence; (5) no more than 9 consecutive GC sequences; (6) the 5' end of the sense strand is preferably G / C; (7) antisense The 5' end of the strand is preferably A / U; (8) the 15-19 bases of the sense strand preferably contain more than 3 A / U; (9) the 5' end of the antisense strand is best among the 7 bases There are more than 5 A / U.
[0033] Use Blast (www.ncbi.nlm.nig.gov / Blast) to perform homology analysis on the candidate sequence and the genome database to ensure...
Embodiment 2
[0037] Example 2 shRNA design and synthesis
[0038] Apply the shRNA design principles (Sun G, Molecular Properties, Functional Mechanisms, and Applications of Sliced siRNA. Mol Ther Nucleic Acids. 2015 Jan 20; 4:e221.), according to the above siRNA-R-Spondin1 sequence, design the shRNA sequence: (1) The two complementary oligonucleotides must have restriction enzyme sites at both ends; (2) a C is immediately below the enzyme site to ensure transcription; (3) the shRNA target sequence starts with a G base, To ensure RNA polymerase transcription; (3) The loop adapter sequence inserted by shRNA should be close to the middle of the oligonucleotide, and 5`-TCAAGAG-3` is the most effective; (4) shRNA can only have a specific and unique loop structure;( 5) Insert 5-6 Ts at the end of the shRNA fragment to ensure that RNA polymerase III terminates transcription; (6) G / C content is 40% to 50%; (7) shRNA sequence should avoid more than three consecutive G / C / A / T appears. The obt...
Embodiment 3
[0043] Example 3 Plasmid vector construction
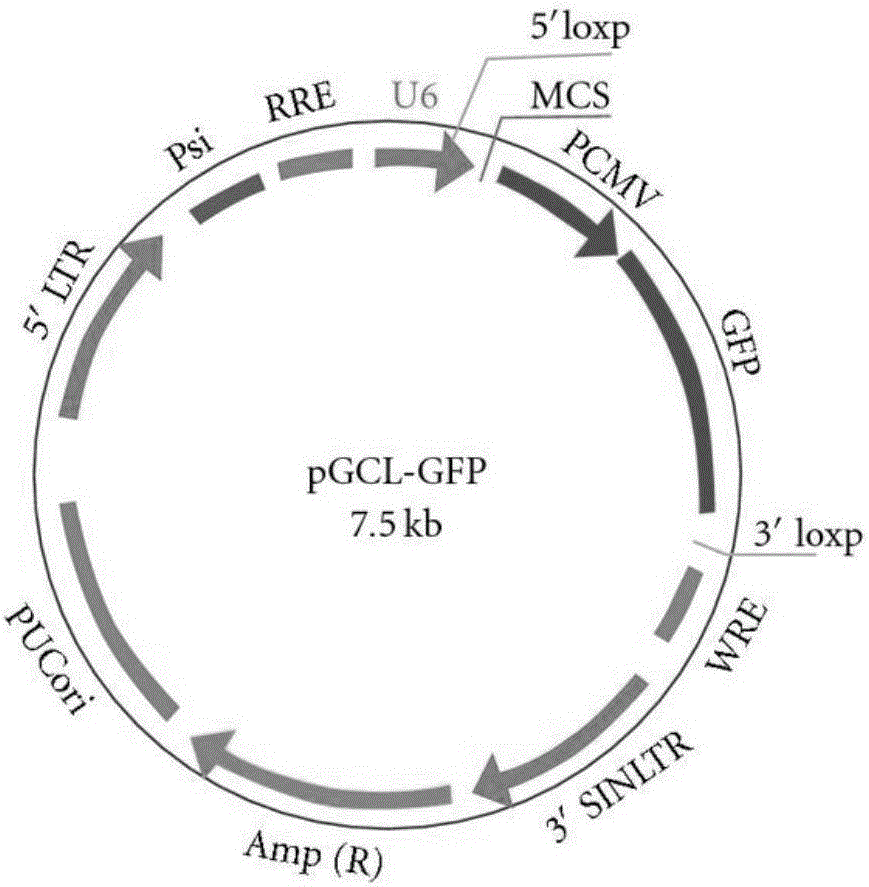
[0044] The above shRNA-R-Spondin1 was synthesized into an oligo DNA single-stranded fragment and annealed to form a double-stranded DNA, which was connected to the pGCL-GFP vector (Shanghai Jikai) to construct the plasmid vector pGCL-GFP-shRNA-R-Spondin1 (see figure 1 ). The plasmid vector will transcribe shRNA in the cell, and produce siRNA-R-Spondin1 targeting the R-Spondin1 gene target after intracellular modification. The annealing and connection process is as follows:
[0045] 1. Annealing into double-stranded DNA
[0046] 1) Establish the following annealing reaction system (room temperature) in a 0.5ml sterile centrifuge tube:
[0047]
[0048] 2) Incubate at 95°C for 4 minutes and at 70°C for 10 minutes;
[0049] 3) Take out the centrifuge tube, place it at room temperature for 5-10 minutes, and cool to room temperature;
[0050] 4) Briefly centrifuge and mix well.
[0051] 2. Ligation of double-stranded DNA to...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com