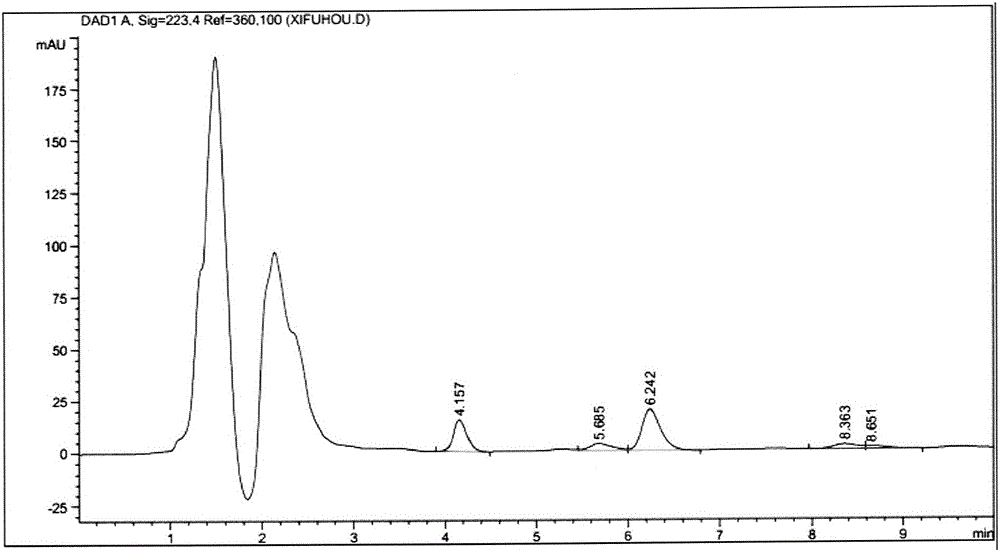
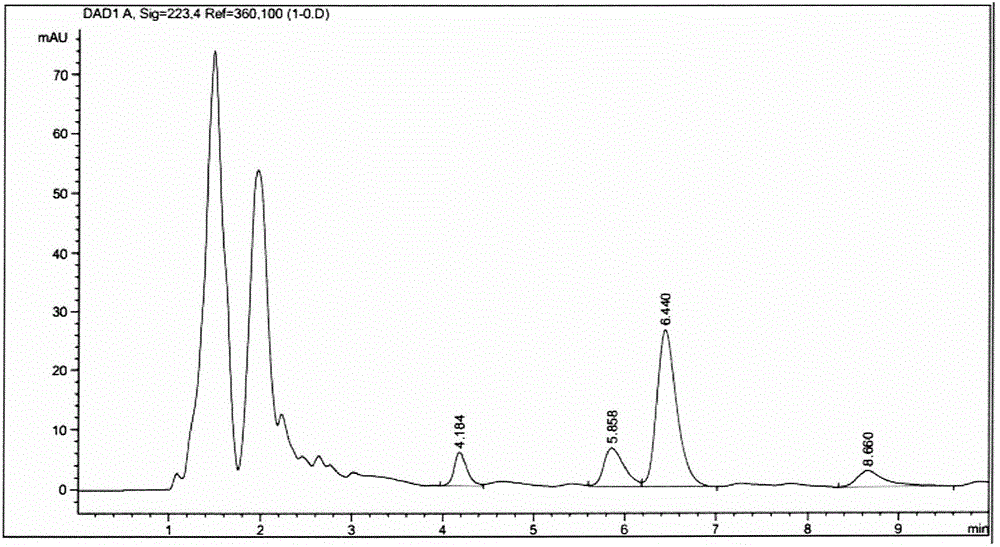
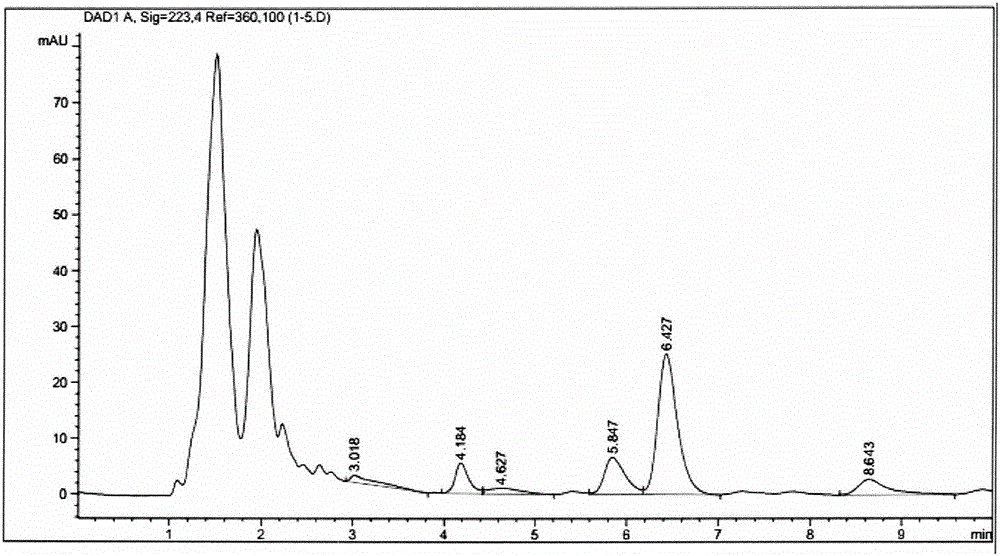
Mutants of fatty acid adenosine monophosphate (AMP) ligase with improved substrate affinities and application thereof
A mutant, ligase technology, applied in the field of genetic engineering and enzyme engineering, can solve the problems of long cycle and high randomness
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] Example 1: Design of mutant genes
[0024] Using computer simulation method, using MOE software for homology modeling, molecular docking of fatty acid AMP ligase and decanoic acid, simulated determination of its affinity, and replacement of some active centers or nearby amino acids that may affect the affinity. For the size of affinity, select sites 296 and 302 with improved affinity.
Embodiment 2
[0025] Embodiment 2: the acquisition of mutant strain
[0026] Adopt the method of PCR amplification and OVERLAPPCR, obtain the amino acid sequence such as the fatty acid AMP ligase gene shown in SEQNO.4 and SEQNO.5, then the gene is connected to pET-22b (+) and then transformed into Escherichia coli E.coliDH5α and carried out Amplify, screen and transform into Escherichia coli E.coliBL21 with correct sequencing, and name the transformants with correct screening as EC296 and EC302.
Embodiment 3
[0027] Embodiment 3: the verification of mutant strain
[0028] Seed culture: Inoculate an appropriate amount of recombinant bacteria EC296 and EC302 from a glycerol tube into LB medium (100ug / ml ampicillin) with a liquid volume of 10ml / 50ml, culture overnight at 37°C on a shaker at 220rpm.
[0029] Shake flask fermentation: Inoculate the overnight cultured seed solution with 2% inoculum into LB medium, and when the OD value is 0.6-0.8, add IPTG with a final concentration of 0.2mM for induction, and induce at 18°C for 18-24h.
[0030] Purification of fatty acid AMP ligase: centrifuge the recombinant bacterial fermentation broth at 8000r / min for 20min, take the precipitate, stir with hypertonic sucrose solution for 20min at low temperature, centrifuge at 12000r / min for 30min, collect the supernatant, and stir the precipitate with hypotonic solution for 20min at 12000rr / min centrifuged for 30min, and the supernatant was collected. The supernatants collected twice were combin...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com