Nucleic acid signal amplification sequence and amplification method
A signal amplification and sequence technology, applied in the field of molecular biology, can solve the problems of increased management costs, false positives, aerosol pollution, etc., and achieve the effects of short detection process, high repeatability and good stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0042] The present invention also provides a method for preparing the above-mentioned nucleic acid signal amplification sequence, which specifically includes the following steps:
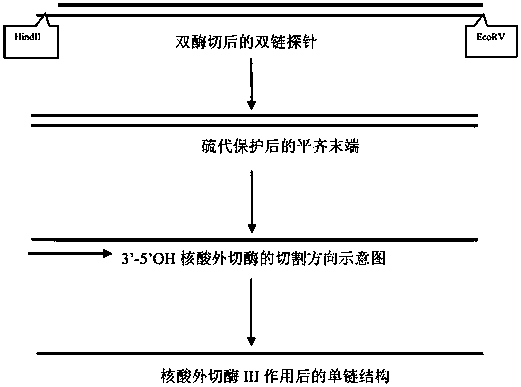
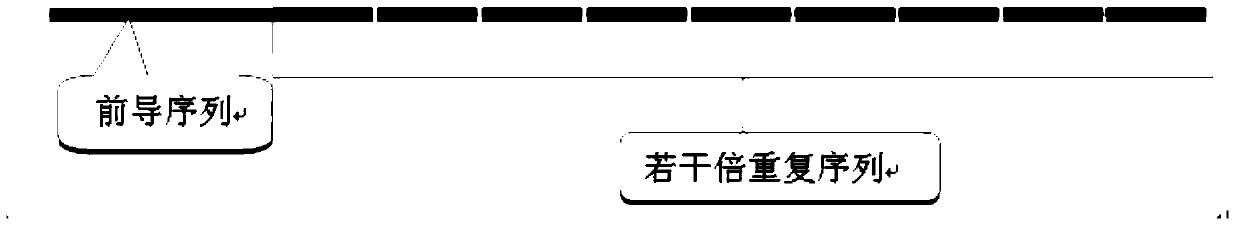
[0043] A. Design a nucleic acid signal leader sequence and multiple repeat sequences according to the target target, and add a restriction endonuclease I cutting site at the 5' end of the leader sequence, and add a restriction endonuclease I at the 3' end of several multiple repeat sequences The enzyme cutting site of restriction endonuclease II; the enzyme cutting site of the restriction endonuclease I is connected with the leading sequence of the nucleic acid signal amplification sequence, and becomes a cohesive end after enzyme cutting; the restriction endonuclease II The enzyme cutting site is connected to the repeat sequence of the nucleic acid signal amplification sequence, and the end is blunt after enzyme cutting;
[0044] B. Cloning and replication after the synthetic sequence is inserted i...
Embodiment 1
[0064] Embodiment 1, the preparation of nucleic acid signal amplification sequence
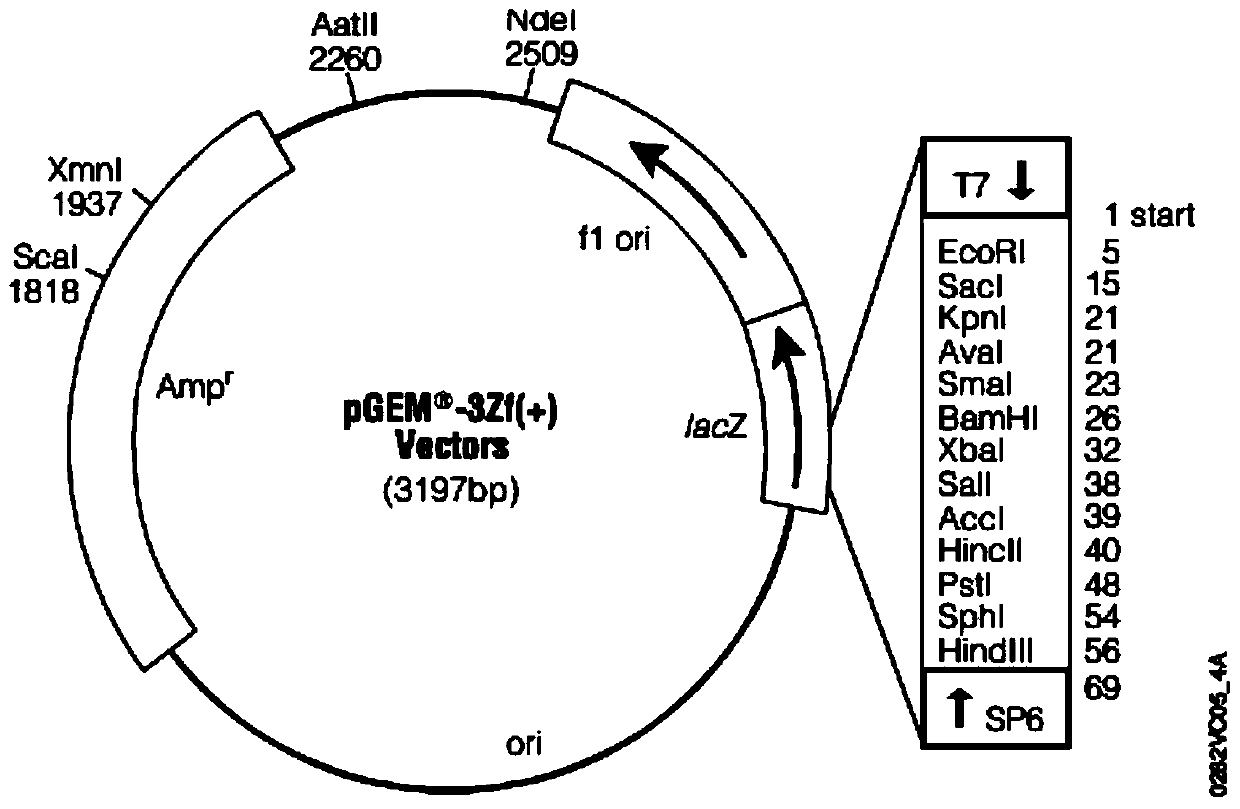
[0065] Design the leader sequence according to the primer sequence 1, and design the repeat sequence of different multiples at the same time, and add the HindII enzyme cutting site at the 5' end of the leader sequence, and the EcoR V enzyme cutting site at the 3' end of the repeat sequence, and synthesize After different conformational sequences were inserted into the pGEM-3ZF(-) vector, clone and replicate; HindII single-digested the vector sequence to generate 5' sticky ends, and used Kelnow large fragment polymerase and adenosine thiotriphosphate to fill in the 5' sticky ends, and then used EcoR V excises the target band from the vector, and exonuclease III enzyme cuts the single-stranded DNA probe. Each sequence is shown in Table 1.
[0066] Table 1 each sequence
[0067] sequence name sequence target ATCGTTAGGCATTCAGGGA leader sequence TCCCTGAATGCCTAACGAT ...
Embodiment 2
[0068] Embodiment 2, primary signal amplification
[0069] 1. Coat the target target in Table 1 with a concentration of 100pmol on a microwell plate;
[0070] 2. According to the method of Example 1, each sequence was synthesized and prepared into a single-stranded probe, and the probes prepared by Synthetic Sequence 1, Synthetic Sequence 2, and Synthetic Sequence 3 were respectively named as pre-amplification 1, pre-amplification 2, and pre-amplification 3;
[0071] 3. Control the hybridization temperature between 50°C and 55°C, add pre-amplification 1, 2, and 3 to each well at a concentration of 100 pmol / μL, and hybridize for 40 to 50 minutes;
[0072] 4. Add the reporter probe (AAAAGAATATTGGTGGTAAAGACCT-AP) respectively, and react at 51℃~53℃ for 30~40min;
[0073] 5. Add the chemiluminescent substrate lumi-Phos Plus (Beckman Inc.), and read the experimental results.
[0074] The layout design is shown in Table 2, and the corresponding data results are shown in Table 3.
...
PUM
| Property | Measurement | Unit |
|---|---|---|
| melting point | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com