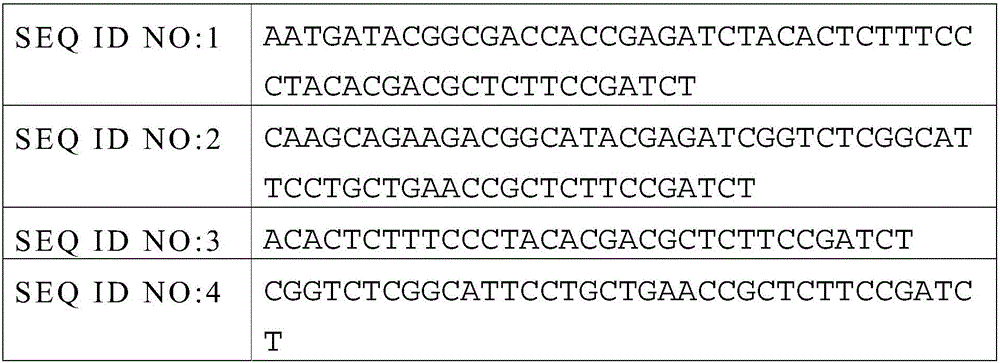Kit and application thereof, and method and system for detecting area target variation
A target area and kit technology, applied in the field of biomedicine, can solve the problems of fewer detection sites, long detection time, cross-contamination of samples, etc., and achieve high specificity, low false positive rate, high cost performance and targeted effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0060] Embodiment one design chip
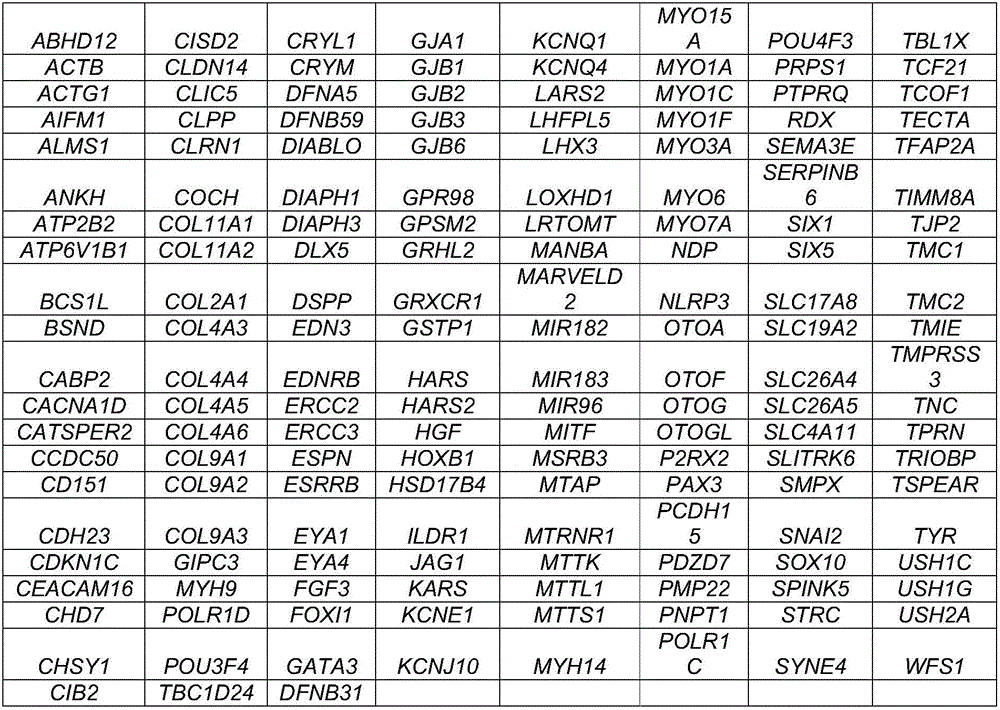
[0061] Both chromosomal variation and single gene can lead to hereditary deafness, and chromosomal variation includes changes in chromosome number and structure, uniparental diploidy and chimerism can also lead to hereditary deafness. The present invention is only aimed at hereditary deafness caused by a single gene. Through the search of the OMIM database and related literature, there are currently more than 400 monogenic diseases related to hereditary deafness caused by 163 single genes.
[0062] According to the human genome HG19, the exon sequences of 163 genes and the flanking ±10-20bp regions were selected for capture sequencing, and the chip size was about 1.6M. The chip is covered with abundant capture probes, with a probe coverage area of 99.0%, which can enrich target DNA fragments from complex genomes, and capture about 1.6M DNA fragments with high specificity and high coverage on the same chip genomic region. The genes are d...
Embodiment 2
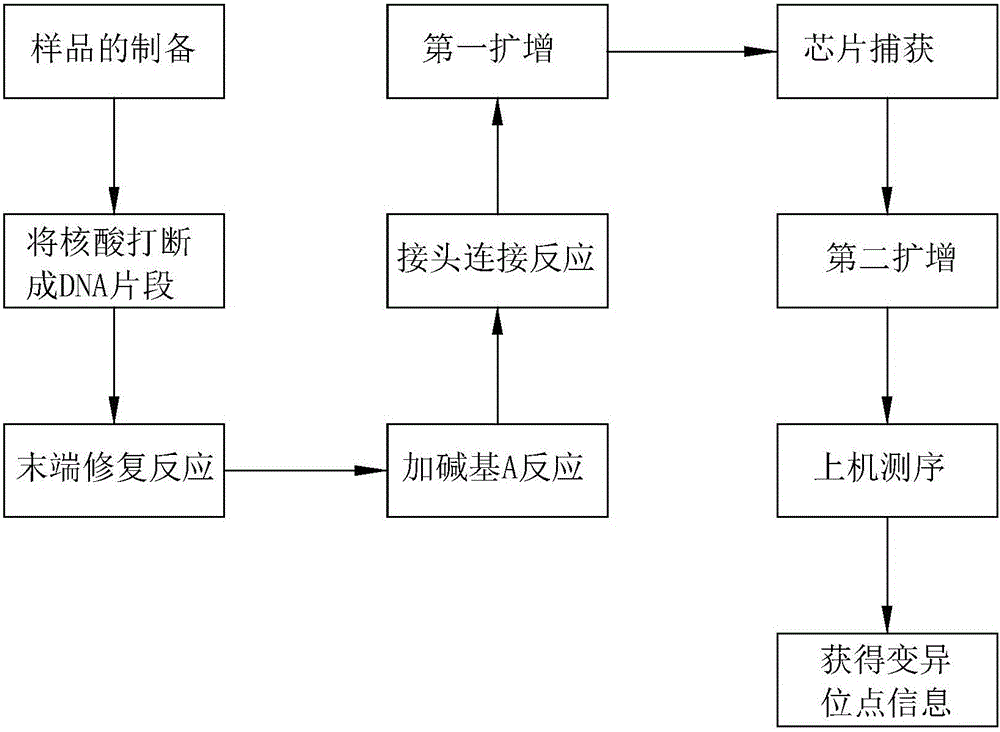
[0063] Example 2 Detection of variation in the target region, for specific procedures, see figure 1 .
[0064] (1) Sample preparation
[0065] Extract sample DNA with conventional DNA extraction methods, especially the salting-out method. Ultrasonic fragmentation of large fragments of DNA, the current sample fragmentation method is the Covaris fragmentation method, which fragments the sample DNA into fragments in the range of 100-700bp. Preferably in the range of 200-250bp.
[0066] (2) Library construction
[0067] 1. End repair
[0068]
[0069] After the reaction, add 180 μL of Ampure Beads, after the magnetic beads are purified, and finally redissolve 30 μL of ddH 2 O, carry out the next reaction with magnetic beads;
[0070] 2. Add A at the end
[0071]
[0072] 3. Joint connection
[0073]
[0074] After the reaction, add 75 μL of Ampure Beads for magnetic bead purification, using 35 μL of ddH 2 O back to dissolve;
[0075] 4.Non-Captured sample Pre-LM...
Embodiment 3
[0084] Example 3 Sequencing Data Analysis
[0085] 1. Obtain sequencing data according to the method in Example 2.
[0086] 2. Off-machine data filtering Reads_filter: filter reads that meet the analysis requirements. Two conditions need to be met: 1) the number of N in the reads is <10%; 2) the bases with a quality value <5 do not exceed 50%.
[0087] 3. Sequence alignment: Bwa aln->sampe|samtools view|samtools sort: compare with the human reference genome sequence to obtain the position and quality information of each reads on the chromosome. The compared files exist in bam format;
[0088] 4. Deduplication MarkDuplicates.jar: Mark the reads aligned to the same starting point of the reference genome as duplicates, and only analyze as one reads in subsequent analysis;
[0089] 5. Re-alignment: GenomeAnalysisTK.jar-T Realigner, TargetCreator, IndelRealigner: use other alignment tools to re-align reads with poor quality in the previous alignment to improve data utilization; ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com