Disulfide-bond-introduced omega-aminotransferase mutant and application thereof
A technology of mutants and transaminases, which is applied in the field of ω-transaminase mutants, can solve problems such as further improvement of thermal stability, and achieve the effect of improving thermal stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
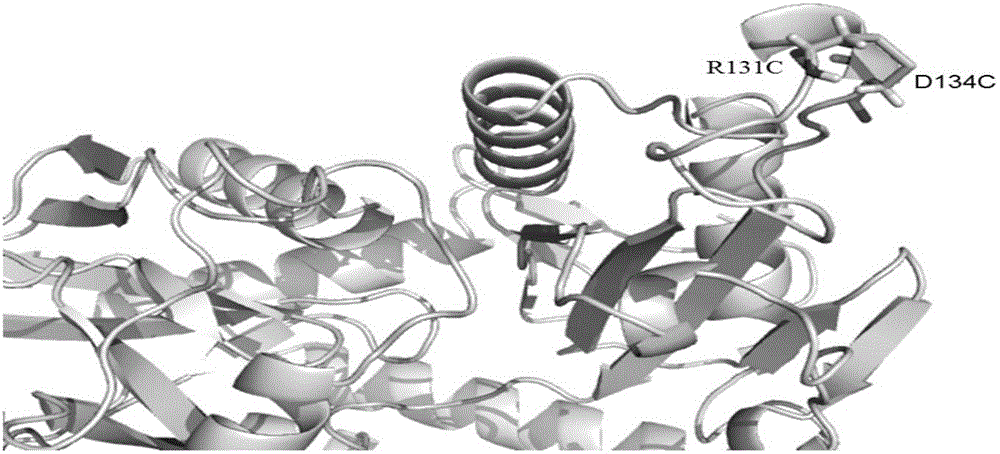
[0029] First upload the PDB file (PDB ID: 4CE5) of Aspergillus terreus ω-transaminase to Disulfideby Design (DbD, http: / / cptweb.cpt.wayne.edu / DbD2) and Disulfide Bonds in Proteins (MODIP, http : / / caps.ncbs.res.in / dsdbase / modip.html) website, the above software fully considered the C-S bond rotation angle of two cysteines, C α distance between and C β Using bioinformatics to calculate and analyze the bond length, bond angle and energy to predict the feasible introduction of 8 disulfide bond sites, they are R131C and D134C, N25C and A28C, D113C and Y146C, E213C and V234C , A44C and L48C, M150C and M280C, A32C and F142C, R161C and Y201C. According to the crystal structure of ω-transaminase, the temperature factor (B-factor) value of amino acid residues in the G129-D134 region is higher. For proteins, if the B-factor value of a certain amino acid residue is higher, it indicates that the The structure where the amino acid residues are located is more unstable. Therefore, in the ...
Embodiment 2
[0031] Site-directed mutagenesis was performed on R131C and D134C of the ω-transaminase gene locus in turn by site-directed mutagenesis PCR. The primers for site-directed mutagenesis are shown in Table 1, thereby obtaining a mutant containing two cysteines at the same time. The mutant was sequenced to verify that its nucleotide sequence was mutated at codons 497 and 499, and the codes of arginine (R, Arg) and aspartic acid (D, Asp) were encoded by positions 131 and 134. The codons CGT and GAT are all mutated to the codon TGC encoding cysteine (C, Cys).
[0032] Table 1 Primers for site-directed mutagenesis.
[0033] Primer name
Sequence 5'→3'
R131C-F
GGTGCGAGGAACTTGCCCGGAAGATATAGTG
R131C-R
CTATATCTTCCGGGCAAGTTCCTCGCACCCC
D134C-F
GGAACTTGCCCGGAATGCATAGTGAACAACCTGTAC
D134C-R
ACAGGTTGTTCACTATGCATTCCGGGCAAGTTCCTC
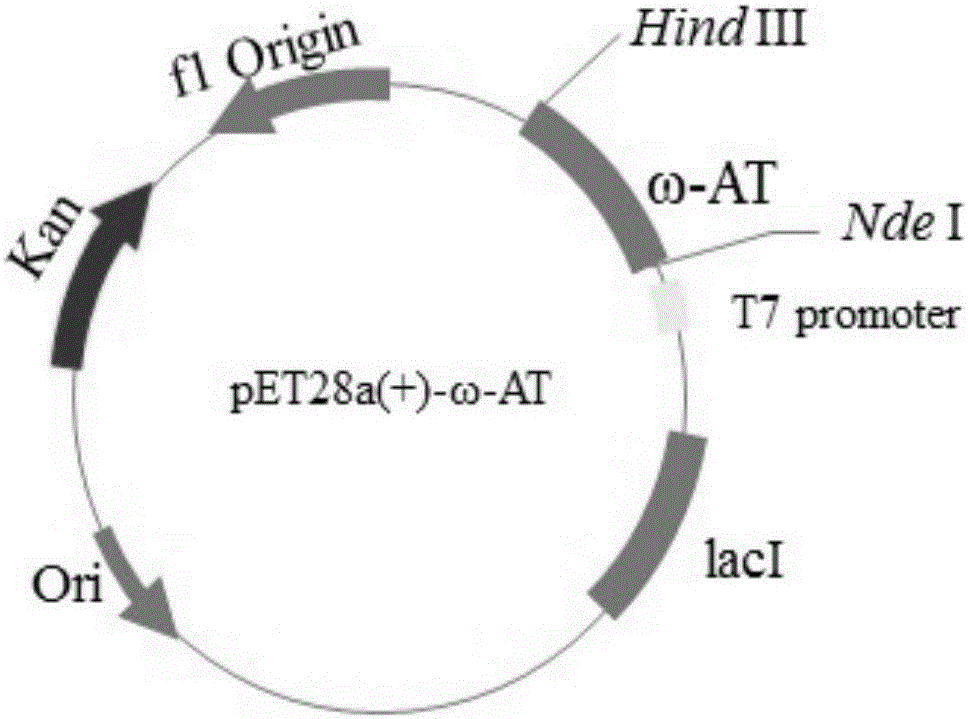
[0034]Using the pET28a-ω-opt-TA plasmid containing the wild-type ω-transaminase gene as a template, t...
Embodiment 3
[0038] Transform the plasmid with the double mutant ω-transaminase mutant gene sequenced correctly in Example 2 into E.coli BL21(DE3), pick a single colony and inoculate it into a test tube with 5 mL of LB liquid medium, at 37°C , 200r / min under the condition of cultivating overnight. Inoculate the 100mL LB medium (10g of tryptone, 5g of yeast powder, 10g of sodium chloride, and adjust the pH of ), cultivated to OD at 37℃, 180r / min 600 When the value is 0.4-0.6, add an appropriate volume of IPTG (final concentration: 0.5mmol / L), and then induce culture at 25°C and 150r / min for 18h, then collect the bacteria.
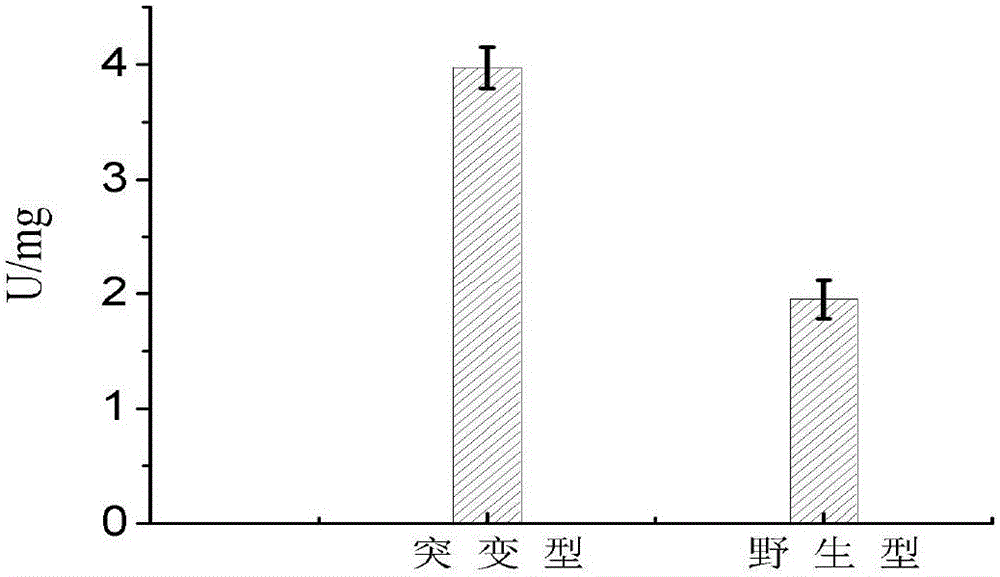
[0039] The collected bacteria were washed twice with phosphate buffer, resuspended in cell-breaking buffer, and ultrasonically disrupted. The working conditions of ultrasonic cell disruption were: power 300W, working for 3s, intermittent for 6s, and ultrasonicating for 8min. The broken cells were centrifuged at 12000r / min and 4°C for 30min, and the supernatant was coll...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com