Compositions and methods for preparing sequencing libraries
A technology for sequencing libraries and sequencing, applied in the field of compositions and methods for preparing sequencing libraries, capable of solving problems such as low coverage of genetic material, sample loss, and impact on sequencing data quality and output
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
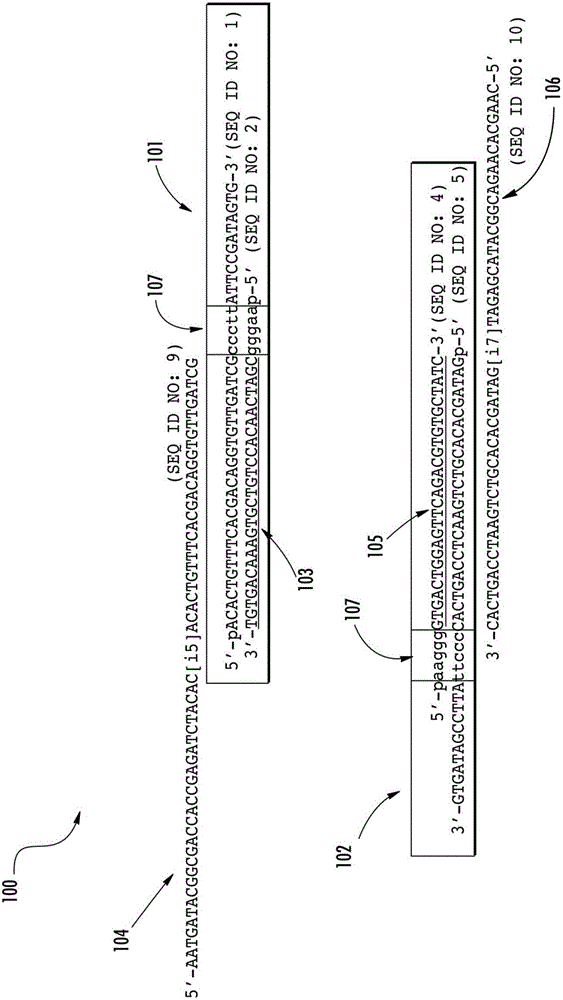
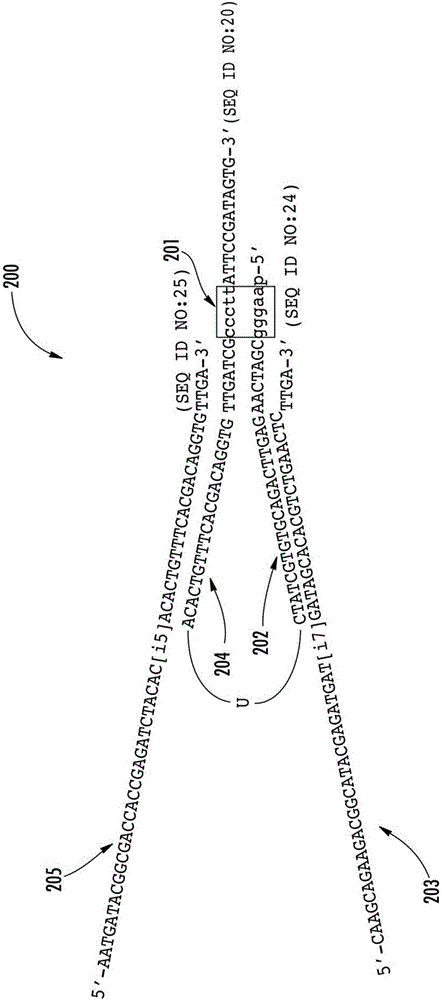
[0134] Preparation of topoisomerase-activated sequencing aptamers
[0135] Activated topoisomerase aptamers (TOPO aptamers) were prepared by hybridizing synthetic oligonucleotides (SEQ ID NO: 1 and 2), and (SEQ ID NO: 4 and 5). The first aptamer of the aptamer set was prepared by hybridizing a first oligonucleotide ACACTGTTTCACGACAGGTGTTGATCCCCTTATTCCGATAGTG (SEQ ID NO: 1 ) to a second oligonucleotide AAGGGCGATCAACACCTGTCGTGAAACAGTGT (SEQ ID NO: 2). The second aptamer of the aptamer set was prepared by hybridizing the third oligonucleotide AAGGGGTGACTGGAGTTCAGACGTGTGCTATC (SEQ ID NO:4) to the fourth oligonucleotide GATAGCACACGTCTGAACTCCAGTCACCCCTTATTCCGATAGTG (SEQ ID NO:5). Hybridization of the oligonucleotides provided a single topoisomerase recognition sequence / site CCCTT (SEQ ID NO: 11 ) for each aptamer. Oligonucleotides (10 μΜ) were hybridized in 10 mM Tris-HCl (pH 7.5), 160 mM NaCl and amplified in a thermal cycler with the following cycles: 98°C for 5 minutes, 85°C for...
Embodiment 2
[0139] Preparation of Sequencing Libraries Using Topoisomerase-Activated Adapters
[0140] To demonstrate the advantages of using TOPO adapters in preparing sequencing libraries for massively parallel sequencing, equal amounts of fragmented sample DNA were used to prepare sequencing libraries according to the topoisomerase-based method provided in this disclosure, and were compared with those using Illumina A ligase-only method to prepare libraries compared to the Illumina method that prepares sequencing libraries in parallel.
[0141] DNA sample preparation
[0142] Lambda DNA or human genomic DNA was sheared into mostly 350bp fragments using a Covaris M220 focused ultrasound generator. Incubate DNA samples (10-500 ng) at 20 °C in a solution containing 10 μl of 5X end repair buffer (1x concentration: 20 mM Tris-acetate, pH 7.9 at 25 °C, 50 mM potassium acetate, 10 mM magnesium acetate, 100 μg / ml bovine serum Protein (BSA)) was end-repaired for 15 minutes in a 50 μl reacti...
Embodiment 3
[0156] DNA sample preparation
[0157] Lambda DNA or human genomic DNA was sheared into mostly 350bp fragments using a Covaris M220 focused ultrasound generator. DNA samples (10-500 ng) were dephosphorylated at 50 °C in a 70 μl reaction mixture containing 10X end repair buffer and 5 μl alkaline phosphatase (shrimp alkaline phosphatase (5 units) or Antarctic phosphatase (25 units)) for 25 minutes. Alkaline phosphatase was heat inactivated at 75 °C for 10 min. Dephosphorylated sample DNA was end-repaired by adding an 8 μl mixture of end repair buffer, dNTPs and 2.5 μl (10 units) of end repair enzymes (Pfu DNA polymerase and / or KOD DNA polymerase) for 5 min at 72°C.
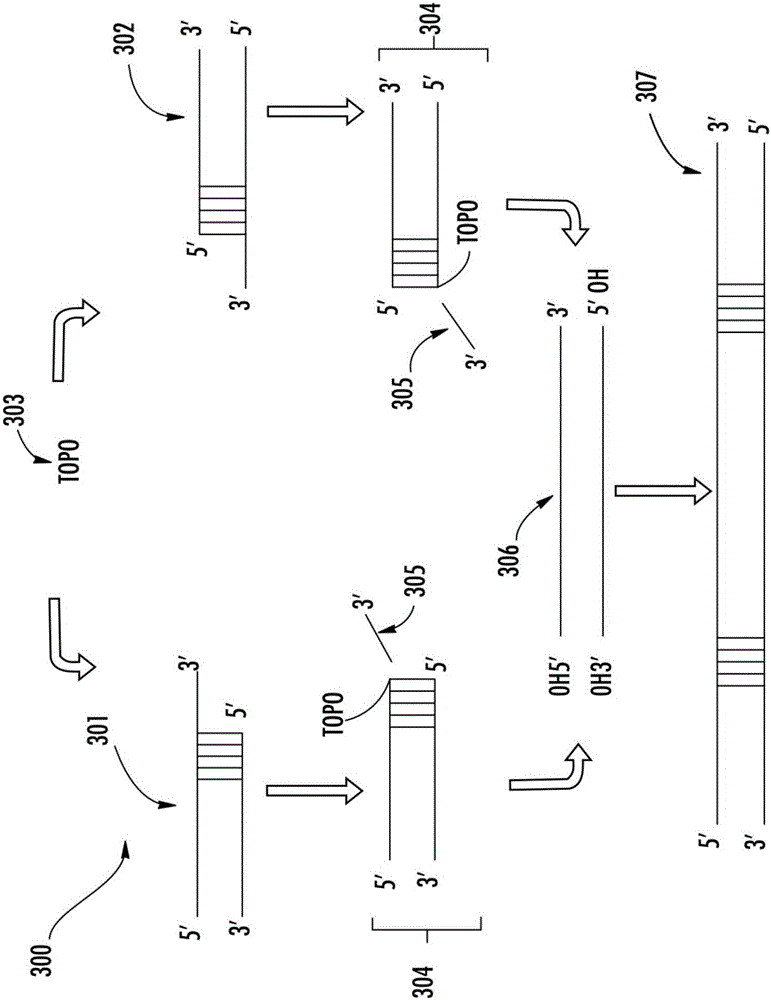
[0158] Topoisomerase-based preparation of sample DNA sequencing libraries
[0159] Dephosphorylation and end repair sample DNA with 2 μl activated topoisomerase aptamer prepared according to Example 1, 2 μl ligase (such as T4 DNA ligase or T7 DNA ligase), 1 μl ATP, 10X ligation in 100 μl reaction mix buffer ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com