Method for detecting 22q11.2 copy number deletion
A copy number and gene copy number technology, applied in the field of molecular biology, to achieve the effect of reasonable program setting, reduced cost input and low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Embodiment 1 detection method
[0034] 1. Primer design:
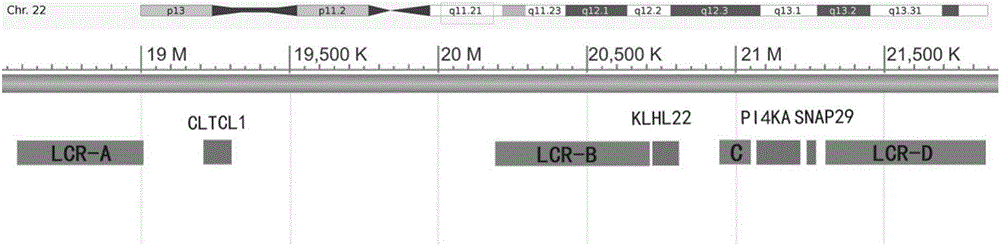
[0035] According to the characteristics of 22q11.2 copy number deletion disease, genes located between different low copy repeats (LCR) in the chromosome 22q11.2 region were selected (LCR22A-B: CLTCL1; LCR22B-C: KLHL22; LCR22C-D: PI4KA / SNAP29) as detection object ( figure 1 ), design four pairs of PCR amplification primers, which are respectively used to amplify fragments in the above genes, and simultaneously select CFTR genes located on different chromosomes, and design primers as reference sequences. When designing primers, the length of the PCR product is controlled at 50-120bps, and the melting curve and Tms value of the product are predicted at the same time, and the Tm difference between the target sequence and the reference sequence is set between 2°C and 10°C. The specificity of the target and reference primers was confirmed by searching the human genome database, and the amplified fragments were kep...
Embodiment 2
[0042] Verification of embodiment 2 detection method
[0043] Material
[0044] Rotor-Gene Q real-time fluorescent quantitative PCR analyzer (Qiagen), KlenTaq enzyme (Ab Peptides), Plus dye (BioFire Defense), primers (BGI Biotechnology Co., Ltd.), human genomic DNA samples (extracted from 22q11.2 microdeletion patients and normal control populations).
[0045] method
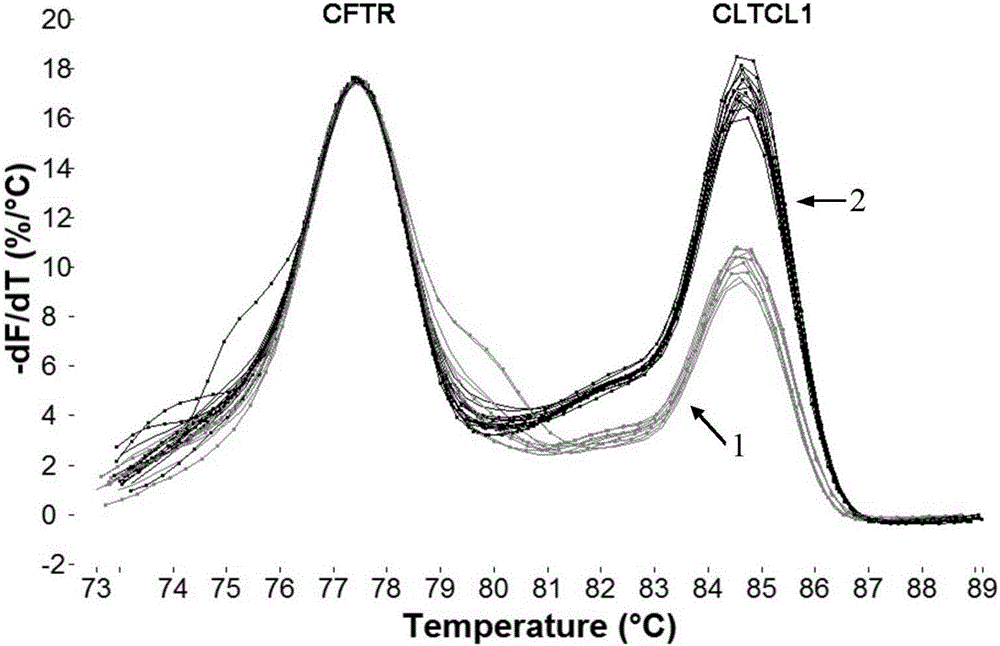
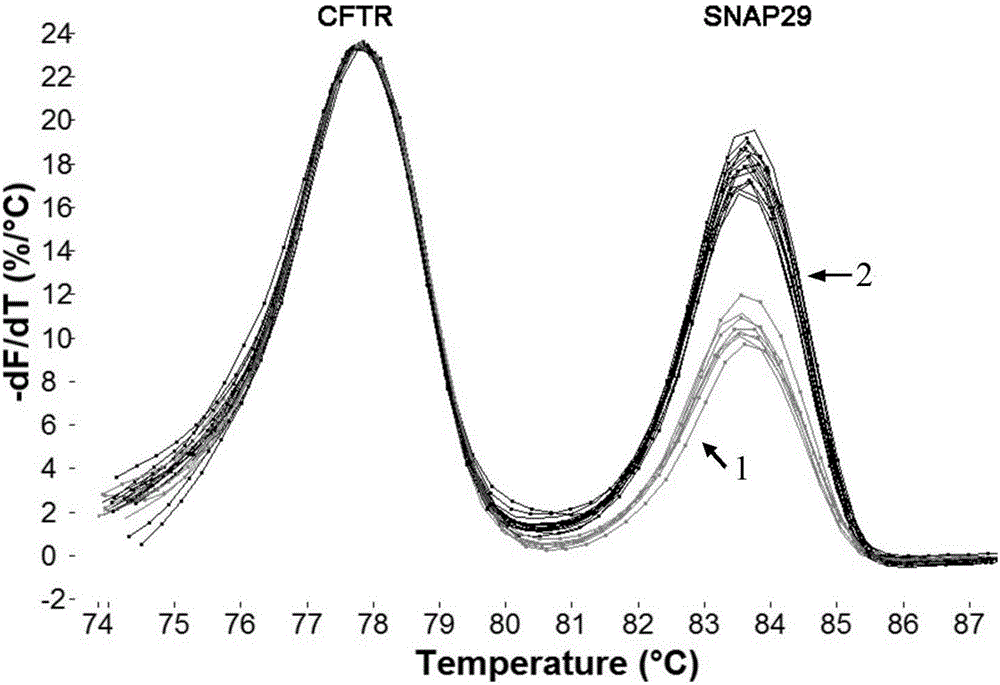
[0046] In order to verify the detection method in Example 1, we used the established system to detect 99 patients with copy number deletion in the 22q11.2 region and normal control samples. Each sample was repeated three times and amplified with multiple junction probes Technology (Multiplex Ligation-dependent Probe Amplification, MLPA) to verify the results.
[0047] result
[0048] In this study, a rapid detection method for 22q11.2 copy number deletion was established by using limited dNTP competitive PCR combined with HRM technology. This system used three competitive PCR reactions to detect four genes ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com