Method of determining cfDNA ratio of donor source in receptor cfDNA sample
A sample-in-receptor technology, applied in the field of biological information and biological detection, can solve the problems of difficult cfdDNA content, inability to be widely used in clinical practice, lack of genetic information, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
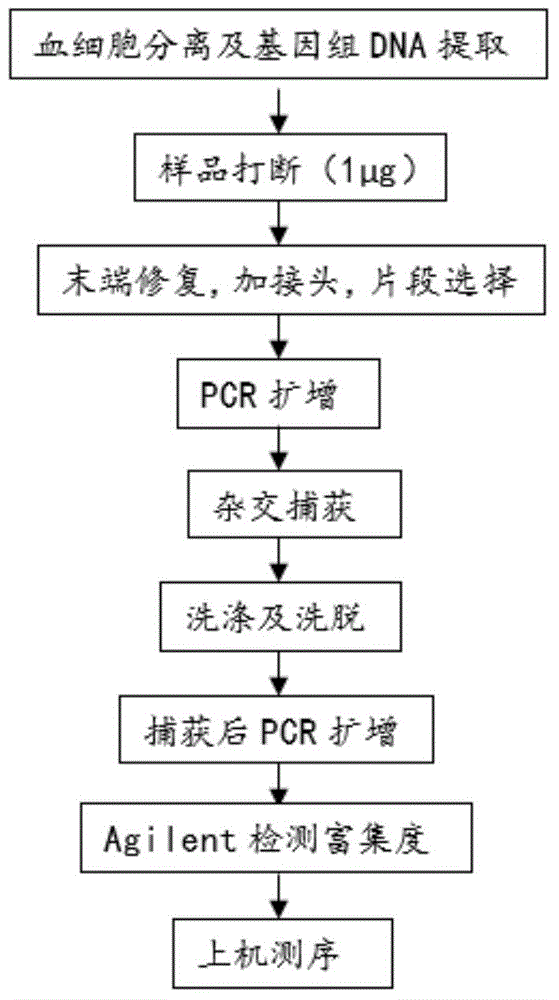
[0065] 1. Experimental methods for obtaining the first sequencing data generally include:
[0066] (1) Target SNP site design
[0067] Since this example method requires a high sequencing depth of SNP sites, which is 200× or more on average, the use of ordinary chips in this method will cause a lot of data waste and greatly increase the detection cost. With the idea that the frequency (MAF) value is closer to 0.5, a small SNP chip was independently designed and synthesized for target area capture.
[0068] The main ways to obtain the target SNP site are as follows:
[0069] 1. The ALFRED allele frequency database is filtered according to the interval of heterozygosity from 0.48 to 0.5 to obtain 946 SNP sites;
[0070] 2. In the 1000Genomes database, filter according to the frequency of 0.5 in the EAS supergroup sub-library, and then filter according to the average frequency of all groups in the library from 0.4 to 0.5, and obtain a total of 2263 SNP sites;
[0071] 3. HapMa...
Embodiment 2
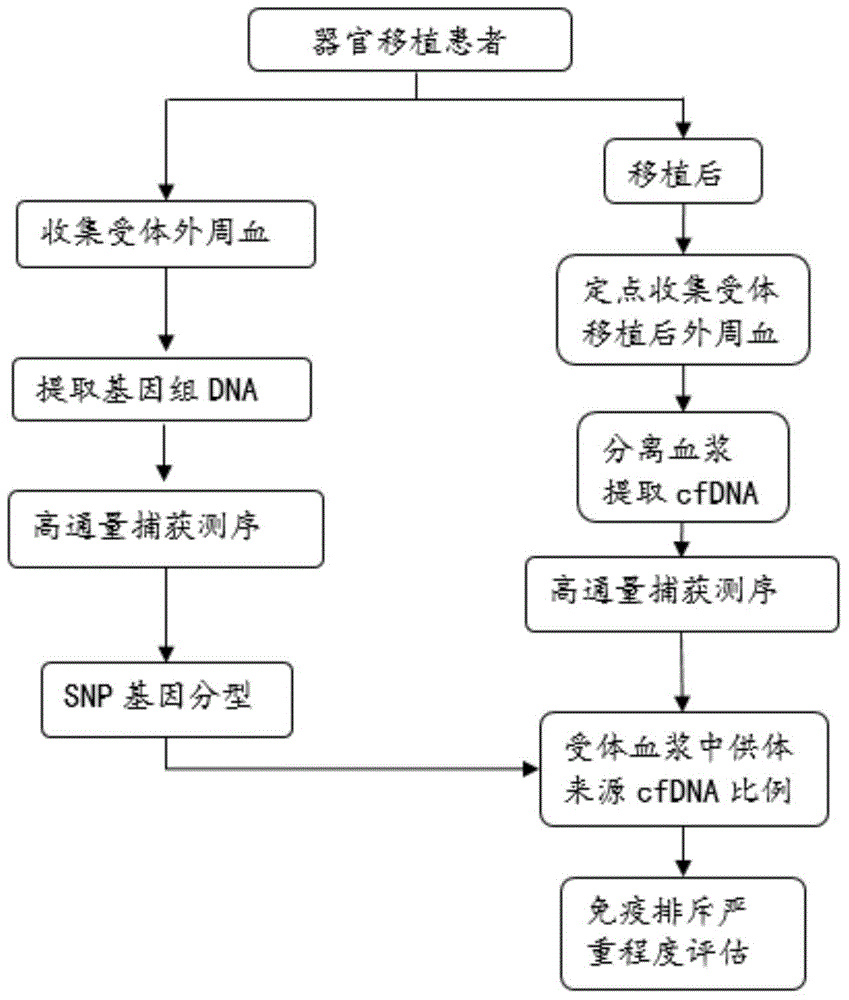
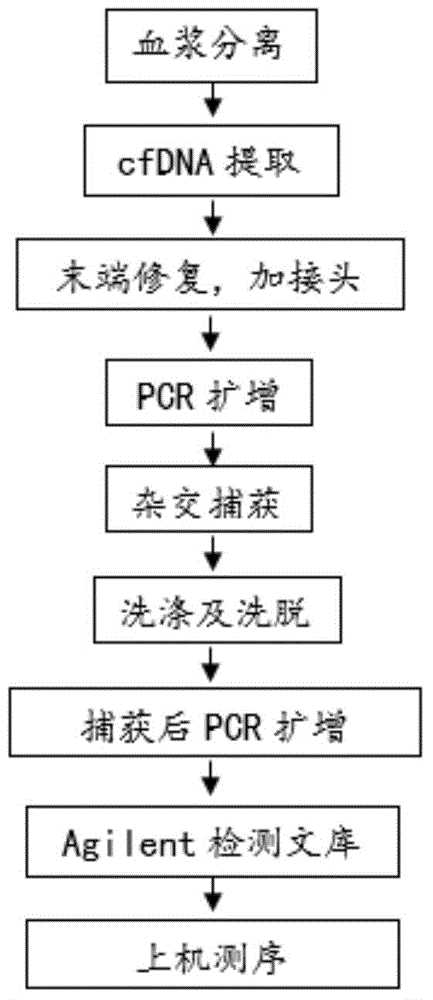
[0128] The design idea of the embodiment is as follows: take 2 normal human blood samples (taken from volunteers), one is the donor and the other is the recipient, mix the samples to be tested, and carry out the simulation experiment. Separation of blood cells and plasma from the blood sample taken, the recipient blood cells (donor blood cells are not required) to extract genomic DNA, interrupt the DNA and perform target region capture sequencing for genotyping; after donor and recipient plasma extract cfDNA, Agelint 2100 The concentration was measured, and the cfDNA of the donor and recipient were artificially mixed according to the ratio of 3.5%, 5.5%, 8%, and 10%, and then the mixed cfDNA was constructed for library capture and sequencing (the sequencer used in this example was the BGISEQ-100 sequencing platform), using To test the reliability of this experimental method. According to the experimental steps in Example 1, the steps in this example are also divided into two...
Embodiment 3
[0323] In order to verify the technical feasibility of using the allele frequency detection method, a simulation verification experiment with known donor ratio was carried out. Taking the BGISEQ-100 sequencing platform as an example, in this embodiment, a normal recipient (sample name R) blood cell sample is selected for target region capture sequencing, and the recipient plasma mixed with donor plasma DNA is also subjected to target region capture sequencing. The proportions were 3.5%, 5.5%, 8%, and 10%, respectively, and the sample names were named according to the mixing ratio. The effective data of the sequencing were compared through tmap comparison, BamDuplicates deduplication, quality control (QC), recipient blood cell genotyping, and recipient blood cell genotyping. The blood plasma frequency statistics and the donor ratio calculation were carried out, and the test reports of the donor content at the 4 blood collection points were finally obtained to evaluate the degree...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com