miRNA related with resistance on phytophthora root rot of soybean and application thereof
A technology of Phytophthora and Phytophthora, which is applied in miRNA and its application fields, can solve problems such as difficulties in the prevention and control of Phytophthora root rot of soybean, achieve good application prospects and reduce resistance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
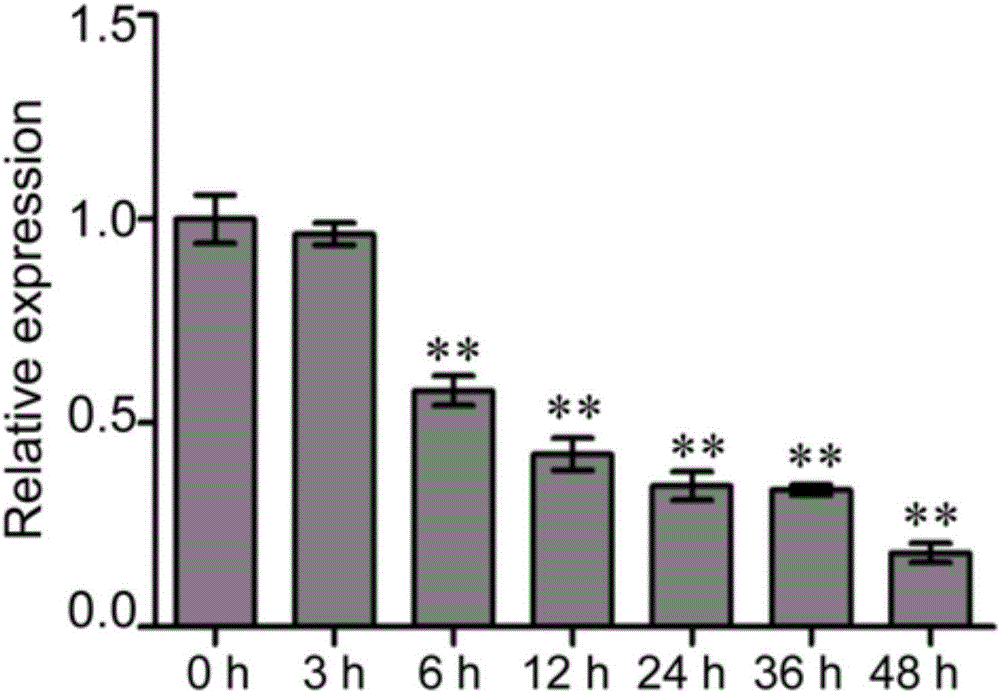
[0022] Example 1. Stem-loop real-time fluorescent quantitative PCR (Stem-loop qPCR) analysis of the role of gma-miR1510b in the mechanism of soybean resistance to Phytophthora root rot
[0023] 1. Design of primers for gma-miR1510b reverse transcription and Stem-loop quantitative PCR
[0024] According to the sequence of gma-miR1510b mature miRNA and the sequence of soybean internal reference gene——GmSnoR I, the primers were designed as follows:
[0025] (1) gma-miR1510b Stem-loop RT primer:
[0026] GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGGTGGA (SEQ ID NO: 3),
[0027](2) gma-miR1510b forward primer: CTGTGCTGTGTTGTTTTACCTAT (SEQ ID NO: 4),
[0028] (3) gma-miR1510b reverse primer: GTGCAGGGTCCGAGGT (SEQ ID NO: 5),
[0029] (4) GmSnoR I forward primer: GAAGATGAAGAGCTTTGTATATTC (SEQ ID NO: 6),
[0030] (5) GmSnoR I reverse primer: ACTCAGAGAGTTGCTTTCTGTG (SEQ ID NO: 7).
[0031] 2. Material cultivation and soybean Phytophthora infection
[0032] (1) Cultivation of Phyt...
Embodiment 2
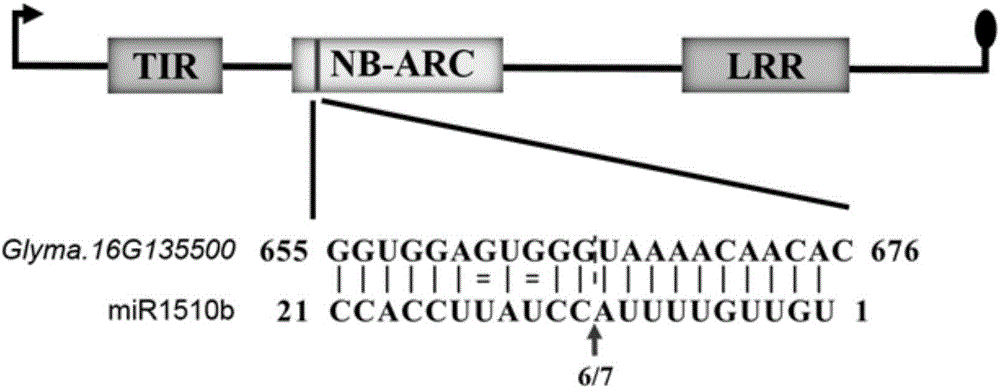
[0061] Example 2. Verification of the gma-miR1510b target gene: use the 5' RACE method to verify the cleavage site of gma-miR1510b to the target gene Glyma.16G135500
[0062] 1. The 5'RACE primers were designed as follows:
[0063] Glyma.16G135500 Outer reverse primer: CCGTGAAGGACATCTCTTCCATTCCAATA (SEQ ID NO:8),
[0064] Glyma.16G135500 Inner reverse primer: CTCCCAATTCATAATCTTTGAAGCAACAAGC (SEQ ID NO:9).
[0065] 2. RNA extraction is the same as in Example 1.
[0066] 3. Enrichment of mRNA:
[0067] (1) Use Promega's mRNA Isolation Systems Kit, for the isolation of mRNA.
[0068] Take 1mg total RNA and add RNase Free dH 2 O dilute to 500 μl; incubate at 65°C for 10 minutes, add 3 μl OligodT, 13 μl 20×SSC, mix gently and cool at room temperature for 10 minutes; take a tube of magnetic beads, flick the bottom of the tube to resuspend the magnetic beads, and place them on the magnetic column Remove the supernatant after 30s; wash the magnetic beads three times with 300μl ...
Embodiment 3
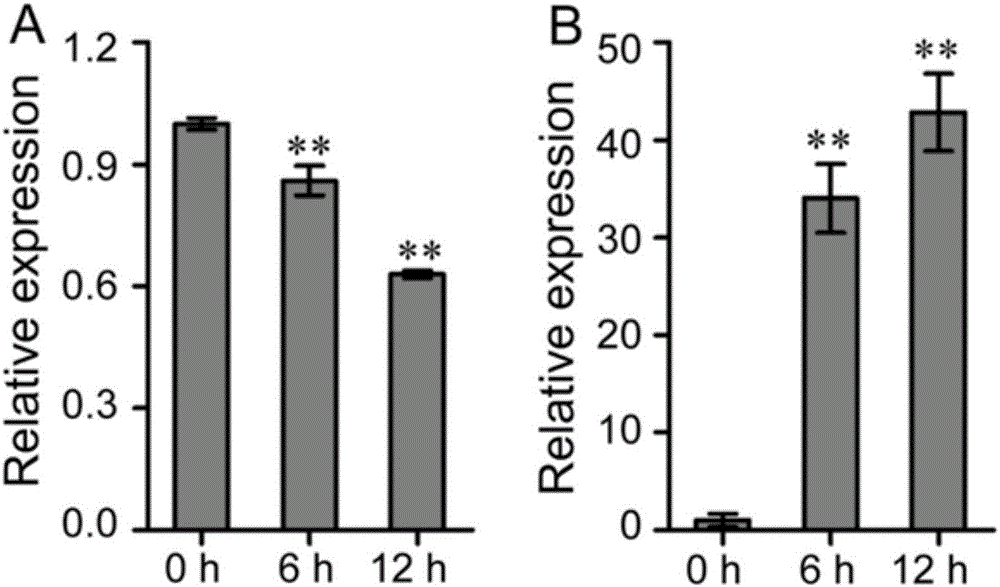
[0082] Example 3. Functional analysis of gma-miR1510b and target gene Glyma.16G135500 in soybean response to Phytophthora soybean
[0083] 1. Planting of soybeans, cultivation of Phytophthora, inoculation method, RNA extraction, reverse transcription and real-time fluorescence quantification refer to Example 1, and the sampling time points are 0, 6 and 12 hours.
[0084] 2. Glyma.16G135500 quantitative primers are designed as follows:
[0085] Glyma.16G135500 forward primer: CTGGTTCCGTAACAAACTCCC (SEQ ID NO: 10)
[0086] Glyma.16G135500 reverse primer: GAAAAACCCCACCAAACAAAGG (SEQ ID NO: 11)
[0087] 3. See image 3 , the results of real-time fluorescent quantitative PCR analysis found that the expression level of gma-miR1510b showed a significant down-regulation trend at 6h and 12h after inoculation with Phytophthora soybean, while the expression level of Glyma.16G135500 was significantly up-regulated at 6h and 12h. After Phytophthora, the expressions of gma-miR1510b and Gl...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com