Biological marker for diagnosing and treating liver cancer
A technology for liver cancer and liver cancer treatment, applied in the field of biomarkers, can solve problems such as functions and mechanisms of action that need to be further explored, and achieve the effect of improving accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
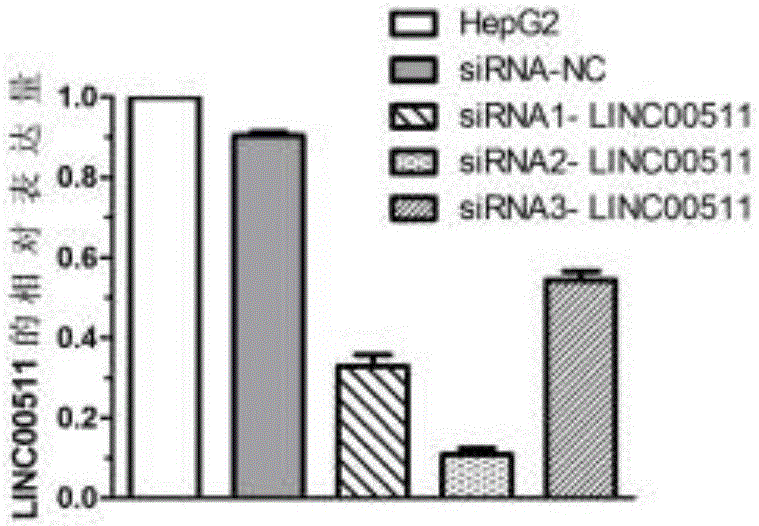
Embodiment 1
[0062] Example 1 Screening Gene Markers Related to Liver Cancer
[0063] 1. Sample collection
[0064] The cancer tissues and paracancerous tissues of 10 patients with liver cancer were collected, and the patients gave informed consent. All the above specimens were obtained with the consent of the organizational ethics committee.
[0065] 2. Preparation of RNA samples
[0066] Tissue RNA was extracted using QIAGEN tissue RNA extraction kit, and the operation was performed according to the specific steps in the manual.
[0067] 3. Reverse transcription and labeling
[0068] The mRNA was reverse-transcribed into cDNA using the Low RNA Input Linear Amplification Kit, and the experimental group and the control group were labeled with Cy3, respectively.
[0069] 4. Hybridization
[0070] The gene chip uses Cannes Bio-Human lncRNA Array, and hybridization is performed according to the steps of the chip instruction manual.
[0071] 5. Data processing
[0072] After hybridizatio...
Embodiment 2
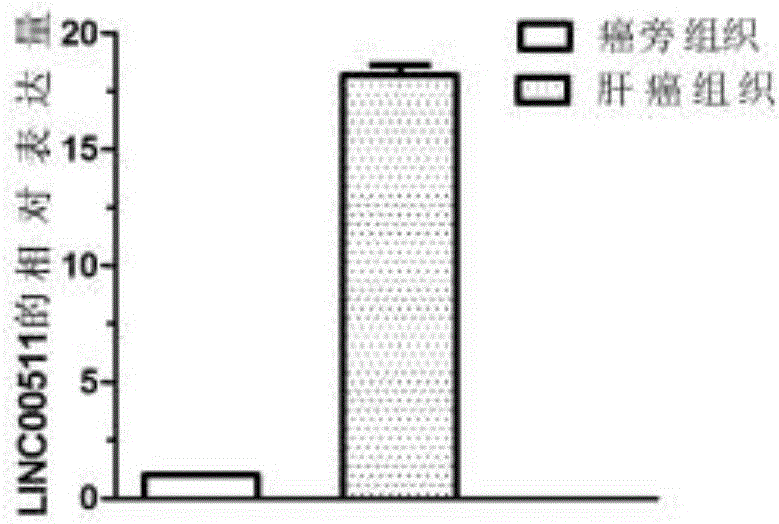
[0075] Example 2 QPCR sequencing to verify the differential expression of LINC00511 gene
[0076] 1. Large-scale QPCR verification of the differential expression of the LINC00511 gene. According to the sample collection method in Example 1, 60 samples of liver cancer tissues and 60 samples of paracancerous tissues were collected.
[0077] 2. The RNA extraction steps are the same as in Example 1.
[0078] 3. Reverse transcription:
[0079] Use 25μl reaction system, take 1μg total RNA for each sample as template RNA, and add the following components to PCR tubes: DEPC water, 5× reverse transcription buffer, 10mM dNTP, 0.1mM DTT, 30μM Oligo dT, 200U / μl M-MLV, template RNA. Incubate at 42°C for 1 hour, then centrifuge briefly at 72°C for 10 minutes.
[0080] (3) QPCR amplification test
[0081] Primer design:
[0082] The primer sequence of LINC00511 gene is:
[0083] Forward primer: 5'-GCTCAAGTTCCTGACATAA-3' (SEQ ID NO.4)
[0084] Reverse primer: 5'-GCGTTGTGTTAGGCATTA-3' ...
Embodiment 3
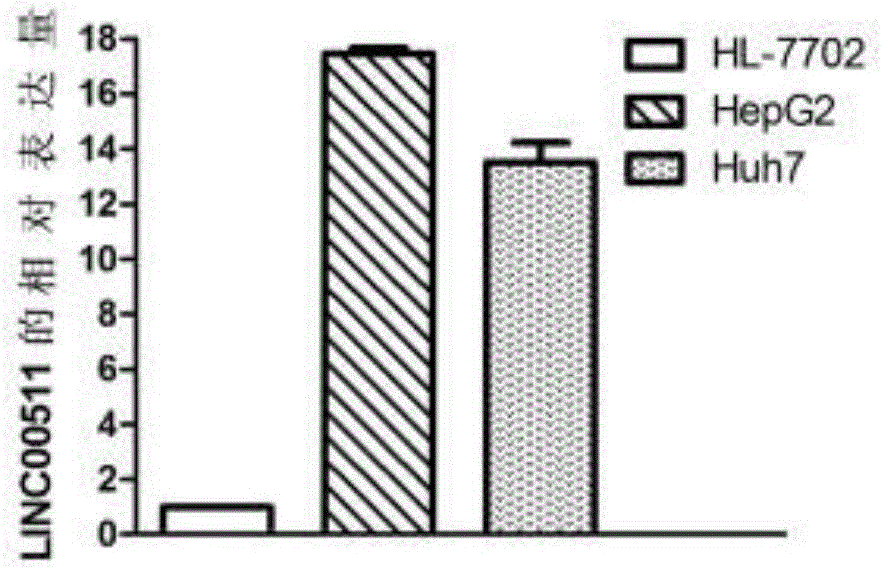
[0094] Example 3 Differential expression of LINC00511 gene in liver cancer cell lines
[0095] 1. Cell culture
[0096] Human liver cancer cell lines HepG2, Huh7 and normal liver cell line HL-7702 were incubated in DMEM containing 10% fetal bovine serum and 1% P / S at 37°C and 5% CO 2 , Cultivated in an incubator with a relative humidity of 90%. Change the medium once every 2-3 days, and use 0.25% EDTA-containing trypsin for routine digestion and passage.
[0097] 2. Extraction of RNA
[0098] 1) Stop the culture when the cells reach 80-90% confluency, digest with 0.25% trypsin and collect the cells in 1.5ml EP tubes, add lm1Trizol to each tube and shake slowly to break the cells, place on ice for 10min.
[0099] 2) Remove protein and DNA: add 0.2ml chloroform to each 1.5ml EP tube, shake for 15s, and place at room temperature for 10min. Centrifuge at 12000rpm for 15min at 4°C.
[0100] The rest of the operation steps are the same as the RNA extraction process from the tis...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com