Primers for detecting phytophthora cinnamomi and nested PCR detection method
A technology of Phytophthora camphora and Phytophthora, which is applied in the field of crop disease detection, identification and prevention, can solve the problems of low accuracy, cumbersome procedures, and long time consumption, and achieve the effects of high accuracy, good repeatability, and easy operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
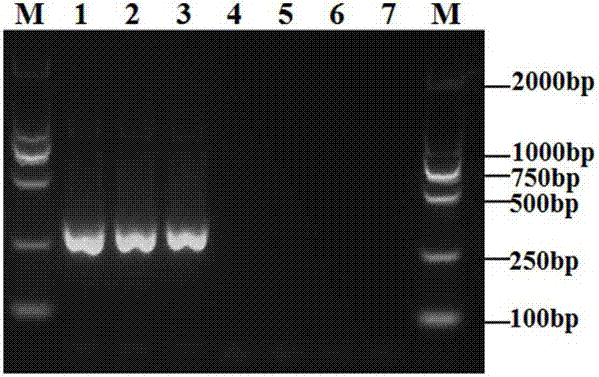
[0029] Example 1: Design of primer pairs for PCR detection of Phytophthora camphora and verification of specificity of primer pairs
[0030] 1. Extraction of Genomic DNA from Phytophthora camphora
[0031] The CTAB method was used to extract the genomic DNA of Phytophthora camphora from different regions. The specific method was as follows: Take a small amount of mycelium powder in a 1.5 mL centrifuge tube (it is better that the mycelium powder just covers the bottom of the semicircle), add 900 µL of 2% CTAB ( cetyltrimethylammonium bromide) extract (2% CTAB; 100 mmol / L Tris-HCl, pH 8.0; 20 mmol / L EDTA, pH 8.0; 1.4mol / L NaCl) and 90 µL SDS ( Sodium dodecylbenzenesulfonate) [Note: CTAB and SDS need to be preheated at 60°C], use a shaker to shake and mix, 60°C water bath for 1h (DNA is released into the buffer), 12000 r min -1 Centrifuge for 15 min; take 700 µL of the supernatant, add an equal volume of phenol, chloroform, isoamyl alcohol mixture (volume ratio 25:24:1), shake g...
Embodiment 2
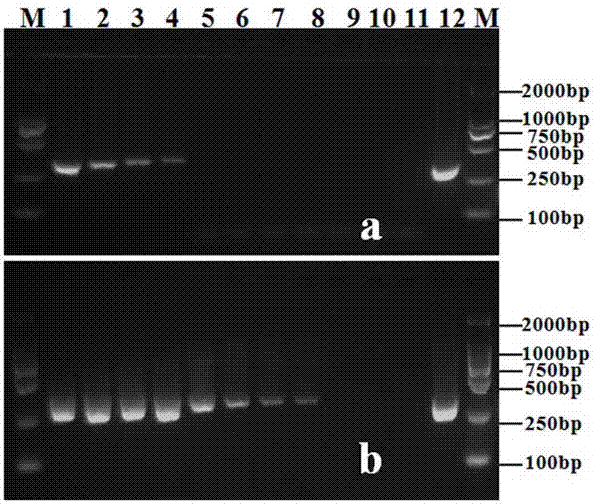
[0040] Embodiment 2: Sensitivity determination of Phytophthora camphora nested PCR detection method
[0041] 1. Conventional PCR amplification
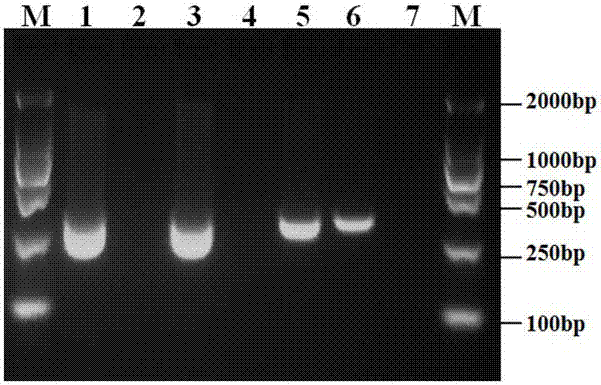
[0042] Genomic DNA of Phytophthora camphora was diluted with sterile ultrapure water, and prepared into series concentrations of 10-fold order of magnitude for future use. Use primer PCINF / PCINR described in the present invention to carry out PCR amplification to the genomic DNA of different serial concentrations, evaluate the sensitivity of this primer to Phytophthora camphora genomic DNA detection, amplification reaction system and reaction procedure are as follows: PCR reaction system 25 μ L, Includes 2 x Taq PCR Master Mix (Beijing Tiangen Biochemical Technology Co., Ltd.) 12.5 µL, 10 µmol / L PCINF / PCINR primers 1.0 µL each, DNA template 1.0 µL, make up to 25 µL with sterile ultrapure water. The amplification reaction program was: pre-denaturation at 94°C for 5 min; 35 cycles of denaturation at 94°C for 1 min, annealing at 59°C...
Embodiment 3
[0048] Example 3: Detection of Phytophthora camphora in the root tissue of blueberry root rot
[0049] Extraction of genomic DNA from diseased root tissue: the diseased root tissue of blueberry root rot caused by Phytophthora camphora infection has been identified by conventional morphology and molecular biology as the sample to be tested, and the DNA is extracted by NaOH rapid lysis method. The specific process As follows: Add 10 µL of 0.5 mol / L NaOH to each mg of plant tissue, thoroughly grind the tissue into a paste in a mortar, transfer it to a 1.5 mL centrifuge tube, centrifuge at 12,000 rpm for 6 min, take 5 µL of the supernatant and add it to 495 µL 0.1 mol / L Tris-HCl (pH=8.0) was mixed evenly, and 1.0 µL was used as a PCR template for amplification.
[0050] Nested PCR amplification detection: using the mentioned DNA as a template, using Ypt1 Gene universal primers ph1F / ph2R (ph1F: 5'-CGACCATTGGCGTGGACTTT-3', Yph2R: 5'-ACGTTC- TCGCAGGCGTATCT-3') for the first round o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com