Efficient zinc ion independent phospholipase C mutant
A technology of phospholipase and lysine, applied in the field of phospholipase C mutants, can solve the problems of shortened shelf life, poor color and taste, and affecting refining effect.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
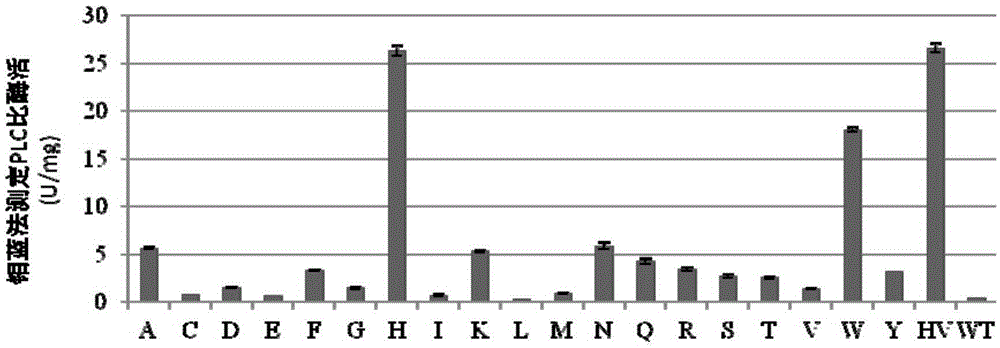
[0142]Example 1: PLC-N63DN131SN134D mutant library construction and screening
[0143] According to the mature peptide sequence (PDB ID: 1AH7) of the phosphatidylcholine-specific phospholipase C of Bacillus cereus and the codon preference of Pichia pastoris, the DNA sequence of BC-PC-PLC (SEQ ID NO: 8, The amino acid sequence encoded by it is shown in SEQ ID NO: 9), and the α factor signal peptide sequence and the Kozak sequence of Pichia pastoris are fused at its front end to finally obtain the α-BC-PC-PLC DNA sequence (SEQ ID NO: 10 ).
[0144] The α-BC-PC-PLC DNA sequence was provided to Shanghai Sangon Biotechnology Co., Ltd. for whole gene synthesis, and the cloning vector pGEM-T-PLC containing the α-BC-PC-PLC DNA sequence was obtained. Using this vector as a template, use The DNA polymerase and the primer pair AmPLC-3 / AmPLC-4 are used to amplify the PLC fragment by PCR.
[0145] Using the pPIC-9k expression vector as a template, use DNA polymerase and primer pair A...
Embodiment 2
[0154] Embodiment 2: PLC-N63DN131SN134D mutant sequence analysis
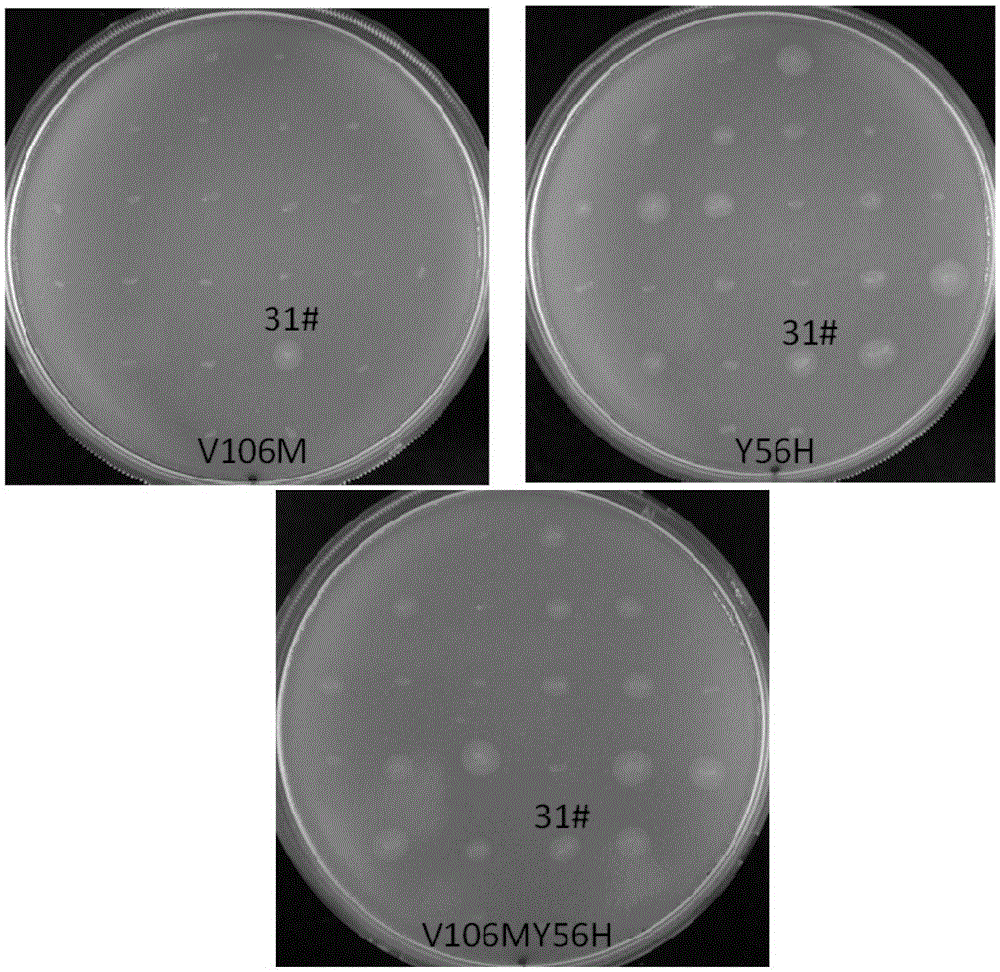
[0155] Inoculate 31# strain in 3ml YPD liquid medium, culture overnight at 30°C, and extract genomic DNA. With the genomic DNA of the 31# strain as a template, use AOX1-5 / AOX1-3 was amplified by DNA polymerase and primers to obtain the DNA sequence of PLC in 31# strain. The obtained sequence was sent to Shanghai Sangon Bioengineering Co., Ltd., and AOX1-5 / AOX1-3 was sequenced with primers. The DNA sequencing results of 31# PLC had 2 base mutations, the 56th tyrosine was mutated to histidine (TAT→CAT); the 106th methionine was mutated to valine (ATG→GTG). The sequence is shown in SEQ ID NO:3.
Embodiment 3
[0156] Example 3: Construction and screening of PLC-N63DN131SN134D mutant single point mutant Pichia pastoris expression strain
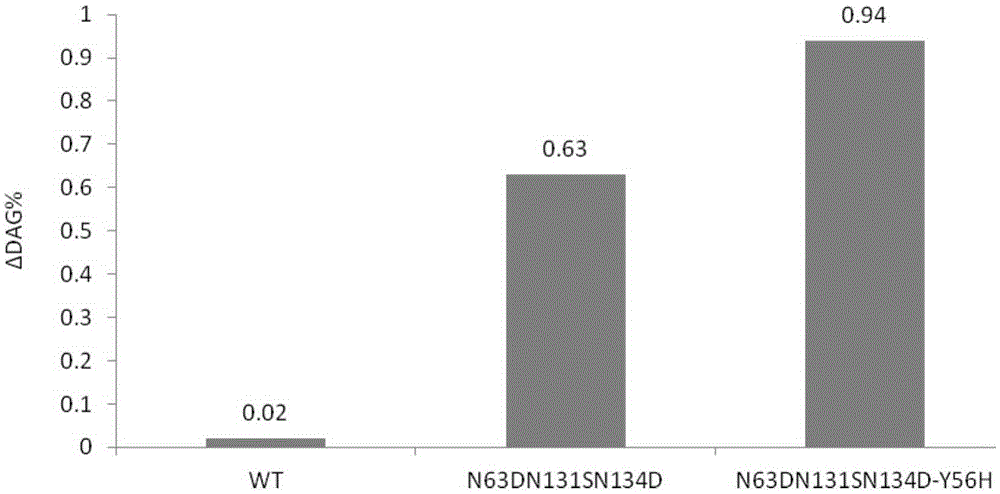
[0157] 1. Construction and screening of PLC-N63DN131SN134D-Y56H
[0158] Using the pmAO-PLC-N63DN131SN134D vector as a template, use DNA polymerase and primer pair EPPLC-1 / 56H-2, a fragment of about 180bp was amplified by PCR. Using the pmAO-PLC-N63DN131SN134D vector as a template, use DNA polymerase and primer pair 56H-3 / EPPLC-2 were used to amplify about 570bp fragment by PCR. The about 180bp fragment and the about 570bp fragment obtained in the previous two steps of PCR were mixed as the template of the third step PCR, and the primer pair EPPLC-1 / EPPLC-2 and DNA polymerase, a fragment of about 755bp was amplified by PCR.
[0159] The about 755bp fragment was cloned into pmAO-PLC through HindIII and EcoRI restriction sites to obtain pmAO-PLC-N63DN131SN134D-Y56H vector. pmAO-PLC-N63DN131SN134D-Y56H was linearized with SalI, and the 8.5kb fr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com