Method for improving trypsin activity through artificially-designed self-activated leading peptide sequence
A trypsin and leader peptide technology, applied in the field of genetic engineering, can solve the problems of low expression, toxicity, and low activation efficiency in vitro
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
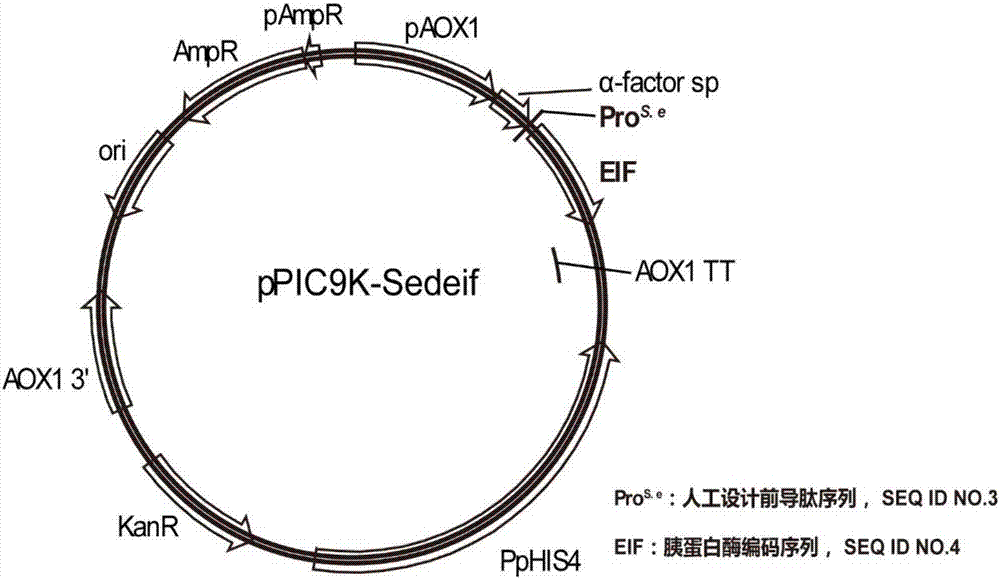
[0023] Example 1 Construction of recombinant plasmid pPIC9K-Sedeif containing recombinant trypsin gene (Sedeif)
[0024] Using the nucleic acid sequence shown in sequence 3 in the patent CN 104328102 A as a template, R primer (sequence shown in SEQ ID NO.8) and F primer1 (sequence shown in SEQ ID NO.9) as primers, perform PCR to obtain a PCR product . Using the PCR product as a template, Rprimer (sequence such as SEG ID NO.8) and F primer2 (sequence such as SEQ ID NO.10) as primers, the nucleotide sequence shown in SEQ ID NO.7 was obtained and named as Sedeif.
[0025] Fusion PCR technology was used to fuse the signal peptide sequence shown in SEQ ID NO.5 at the 5' end of the Sedeif sequence. The fusion fragment and pPIC9K were digested with Not I and BamH I respectively, and purified with T4 ligase overnight at 16°C connect. The ligation product was chemically transformed into JM109 competent cells. The transformation solution was coated with kanamycin (50mg / L) LB plates, ...
Embodiment 2
[0026] Example 2 Construction of high-yielding mature trypsin yeast engineering bacteria
[0027] The Pichia pastoris expression plasmid pPIC9K-Sedeif was linearized with SalI, and the Pichia pastorisGS115 competent cells were transformed by electroporation. The specific method is as follows:
[0028] 1) Inoculate Pichia pastoris GS115 activated on YPD plate in 25mL / 250mL Erlenmeyer flask, and culture overnight at 30°C; inoculate 1% of the above culture solution into 50mL / 500mL Erlenmeyer flask, and the cultured bacteria concentration OD600 is 1.3-1.5;
[0029] 2) Centrifuge at 5000r / min at 4°C for 10min to collect the cells, suspend the cells in 50mL and 25mL of sterile water respectively; 3) resuspend the above cells in 5mL of 1M sorbitol, centrifuge at 5000r / min at 4°C for 10min to collect the cells;
[0030] 4) Resuspend the above cells in 500 μL of 1M sorbitol, aliquot into 80 μL / 1.5mL EP tubes for electrotransformation of competent cells;
[0031] 5) Mix 20 μL of linear...
Embodiment 3
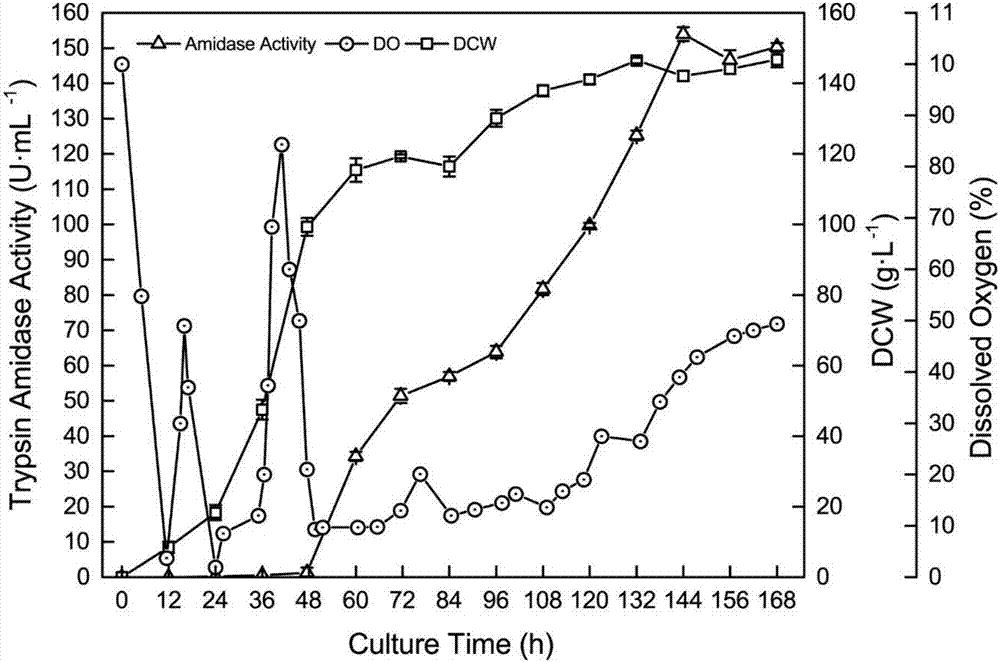
[0035] Example 3 Recombinant Pichia pastoris 3L tank fermentation
[0036] The engineering bacterium constructed in Example 2 is used as the production strain (the recombinant bacterium constructed in the patent CN 104328102 A is used as the control strain), after the activation of the YPD plate, inoculate 50mL / 250mL seed medium, and cultivate it at 30°C for 24h at 220r / min as Fermentation culture seed liquid. 10% was inoculated with 800mL / 3L fermentation medium, pH 5.5, cultured in stages at 30°C: 0-19h, 500rmp / min culture, dissolved oxygen dropped from 100% to about 8%, and then increased to about 60%; 19-34h, The rotational speed gradually increased to 1000rmp / min, and 50% glycerol was fed exponentially, DO began to drop to about 7%, and then rose to 79.1%; 34-168h, 1.8% (V / V) methanol was fed to induce trypsin production.
[0037] Seed medium (g L –1 ): Peptone 20, Yeast Extract 10, Glucose 20.
[0038] Fermentation medium (g L –1 ): Glycerol 40; K 2 SO 4 18; KOH 4.1...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com