Mcr-1 positive bacterium screening method
A screening method and mcr-1 technology, applied in the fields of molecular biology and medical microbiology, can solve the problems of poor plate screening method and low positive rate, and achieve the effect of reducing the workload of PCR
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] 1. Sample Inoculation
[0030] Inoculate a matchhead-sized sample onto an SS plate containing 2ug / ml, and incubate in a 37°C incubator for 18-24 hours. The composition of the SS plate is shown in Table 1.
[0031] Table 1: SS Tablet Composition List
[0032] ingredient name content Peptone (tryptone) 5.0g / L Beef Extract 5.0g / L lactose 10.0g / L Agar 20.0g / L Neutral red 0.025g / L Sodium Thiosulfate 8.5g / L No.3 Bile Salt 3.5g / L Sodium citrate 8.5g / L Ferric citrate 1.0g / L Brilliant Green 0.00033g / L
[0033] 2. Isolation of mcr-1 positive bacteria
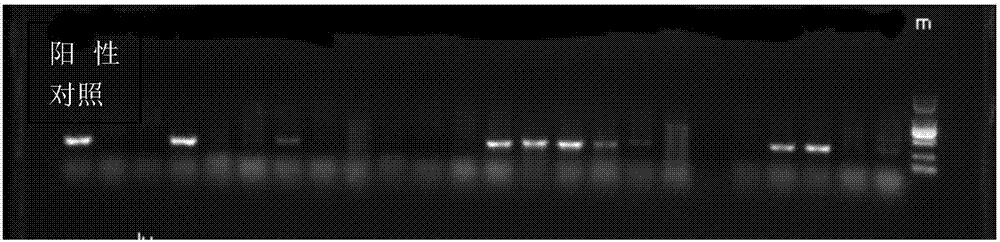
[0034] Pick a single colony on the plate and transfer it to a new plate for purification, and incubate in a 37°C incubator for 18-24 hours; pick the above-mentioned single colony for PCR detection of mcr-1. PCR electrophoresis picture as figure 1 As shown, those with the same band as the positive control are positive for mcr-1; further screen ...
Embodiment 2
[0036] 1. Sample pretreatment
[0037] Inoculate the sample into 5ml of nutrient broth, and incubate the inoculated broth sample in a 37°C incubator for 24-48 hours for testing. The composition of the nutrient broth is shown in Table 2.
[0038] Table 2: Nutrient Broth Composition List
[0039] ingredient name content Peptone (tryptone) 10.0 / L Yeast extract 5g.0 / L NaCl 10.0g / L
[0040] 2. Direct PCR detection of polymyxin resistance gene mcr-1 in broth samples after enrichment:
[0041] Take 1ml of the above enrichment solution into a 1.5ml EP tube, centrifuge at 8000r / min for 3min, remove the supernatant; add 1ml of normal saline to suspend the sediment, wash thoroughly, centrifuge at 8000r / min for 3min, remove the supernatant; add Suspend the sediment in 70ul ultrapure water, boil the suspension in boiling water for 8-10min to free the DNA; centrifuge the suspension at 8000r / min for 3min, and the supernatant is the DNA template for the PC...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com