Micro ribonucleic acid 122 (miR-122) probe and preparation method thereof
A microRNA and probe technology, applied in biochemical equipment and methods, DNA/RNA fragments, recombinant DNA technology, etc., can solve the problems of microRNA function loss and other problems, and achieve low cost, high specificity, and easy operation Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment example 1
[0035] Example 1: Synthesis of small molecules and miR-122 probes
[0036] Step 1: Synthesis of Small Molecules
[0037]
[0038] Dissolve 220 mg or 0.5 mmol of compound 1a in 10 mL of tetrahydrofuran, add 43 mg, 0.5 mmol of compound 1b and 100 μL of DIPEA, respectively, stir overnight, distill under reduced pressure and use directly in the next step.
[0039] Step 2: Synthesis of miR-122 probe
[0040]
[0041] The product obtained in the previous step was dissolved in anhydrous DMSO at a concentration of 5 mM. Amino-modified miR-122 was dissolved (50 μM) in PBS (pH=7.4), and the same volume of DMSO was added to make a 5 mM stock solution. Reacted overnight at room temperature, purified by HPLC and lyophilized to obtain miR-122 probe.
Embodiment example 2
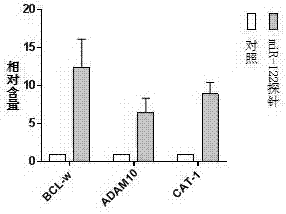
[0042] Example 2: Luciferase experiment:
[0043] Step 1: Construction of reporter plasmid
[0044] The 3'UTR end of the Luciferase reporter plasmid was cut with HindIII and Spel endonucleases, and the resulting fragments were separated and purified by electrophoresis. The complementary sequence of miR-122 was inserted into the purified luciferase reporter plasmid through DNA ligase, T4 ligase, and transformed into competent Escherichia coli, and then screened through a screening plate containing ampicillin to obtain possible positive plasmids. Finally, the reporter plasmid was sequenced to confirm its correctness.
[0045] Step 2: Luciferase Assay Analysis
[0046] The luciferase reporter plasmid with the complementary sequence of miR-122 was transfected into HepG2 cells. First, the HepG2 cells were planted in a 24-well plate. When the cell density reached 70%-80%, 0.25 μg miR-122 The reporter plasmid and 0.15 μg β-galactosidase internal reference plasmid were transfected ...
Embodiment example 3
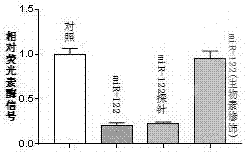
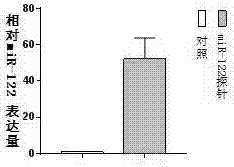
[0047] Example 3: Determination of miRNA expression
[0048] Take out 5 μL of RNA for reverse transcription reaction to prepare corresponding cDNA samples. The specific reverse transcription reaction 10 μL system is:
[0049] 5x AMV buffer (Takara) 2 μL
[0050] AMV (Takara) 0.5 μL
[0051] RT primer (ABI) 0.5 μL
[0052] dNTP mixture (Takara) 1 μL
[0053] DEPC H2O 1 μL
[0054] RNA 5 μL
[0055]The reaction program is: 16°C for 30min, 42°C for 30min, 85°C for 5min, and store at 4°C. The prepared cDNA was then used for real-time quantification of miR-122 by probe method (Taqman) PCR, 20 μL;
[0056] The Taqman PCR reaction system is:
[0057] 10x buffer (Takara) 2 μL
[0058] MgCl2 (25mM) (Takara) 1.2 μL
[0059] Taq (Takara) 0.3 μL
[0060] dNTP mixture (10mM) 0.4 μL
[0061] Probe (ABI) 0.33 μL
[0062] cDNA 1 μL
[0063] Distilled H2O 14.77 μL
[0064] The PCR reaction program was 95°C for 5 min, 95°C for 15s, 60°C for 1min, and 40 cycles of amplification. R...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com