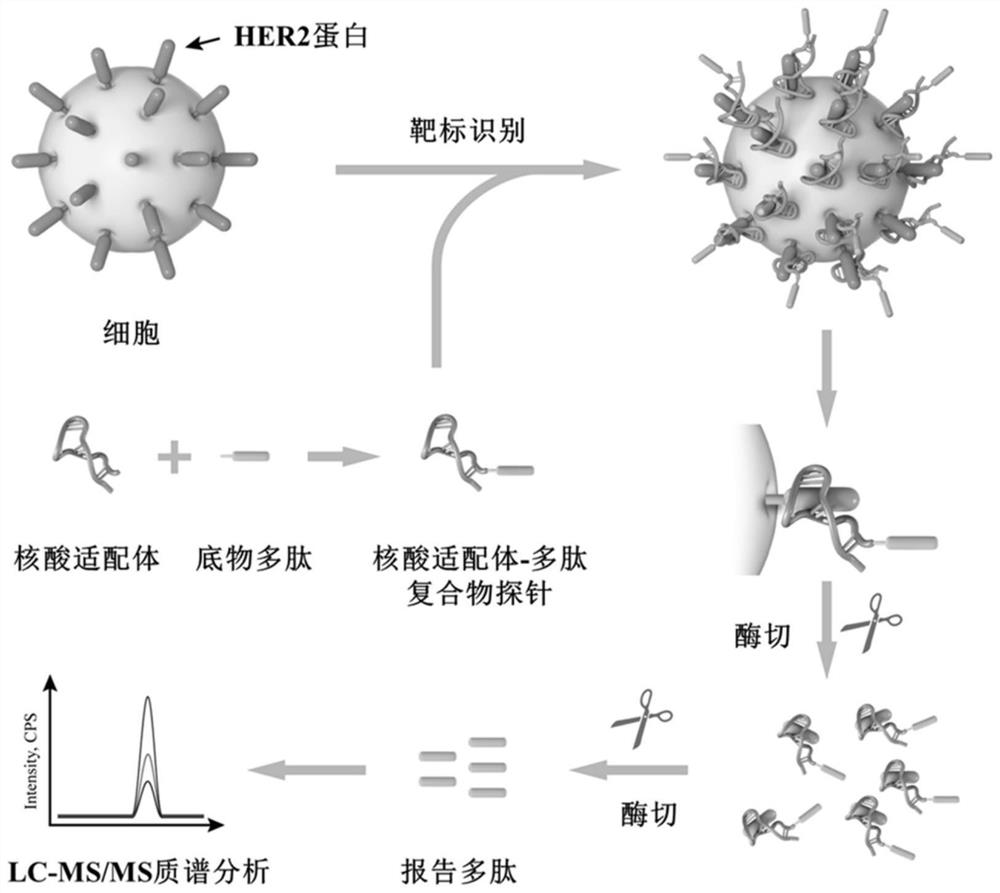
Nucleic acid aptamer-polypeptide complex probe and its preparation method and application
A nucleic acid aptamer and polypeptide complex technology, which is applied in the fields of biology and medicine, can solve problems such as low solubility and low enzymatic hydrolysis efficiency, and achieve high affinity, high purity, and simple and easy preparation methods
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Synthesis and identification of embodiment 1 nucleic acid aptamer-polypeptide complex probe
[0036] (1) Preparation and purification of nucleic acid aptamer-polypeptide complex probes
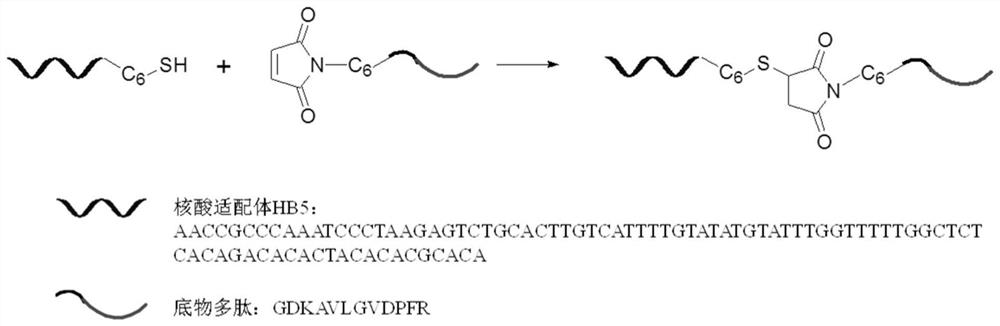
[0037] With trichloroethyl phosphate (TCEP) as reducing agent, 100 μL of disulfide bond-modified nucleic acid aptamer HB5 at a concentration of 20 μM
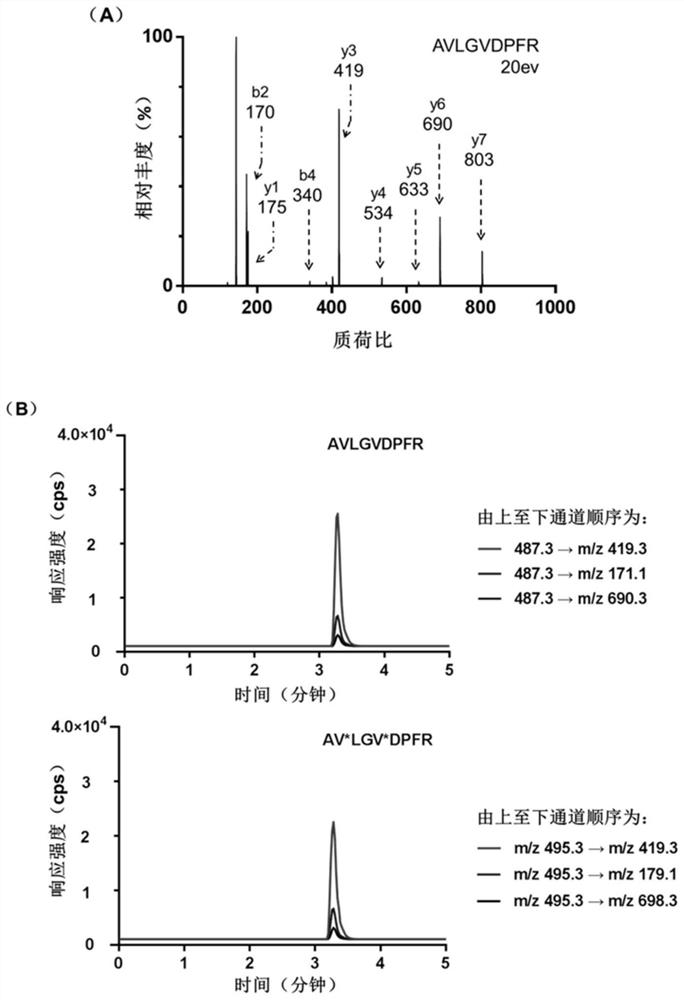
[0038] (AACCGCCCAAATCCCTAAGAGTCTGCACTTGTCATTTTGTATATGTATTTGGTTTTTGGCTCTCACAGACACACTACACACGCACA) was mixed with 50 μL of TCEP-reduced beads, and reacted and shaken at 37°C for 2h. The samples were then centrifuged at 1000 x g for 5 min. Take the supernatant containing reduced HB5, add the same volume of 200 μM maleimide-modified substrate polypeptide (GDKAVLGVDPFR) to the supernatant, shake vigorously at 37°C for 4 hours to carry out the conjugation reaction, and then use conjugation The difference in retention time before and after the yoke (the retention times of the nucleic acid aptamer and the nucleic acid aptamer-polypeptide comple...
Embodiment 2
[0046] Example 2 Detection of HER2 protein content on the surface of breast cancer cells with nucleic acid aptamer-polypeptide complex probe
[0047] In order to prevent damage to the proteins on the cell surface, we collected HER2-positive breast cancer cells (BT474 and SK-BR-3) and HER2-negative breast cancer cells (MDA-MB-231 and MCF -7). After the cells were washed three times with PBS, they were blocked with 200 μL of binding buffer at a blocking temperature of 37° C. for 30 min. Then at a cell number of about 1 x 10 6 200 μL of the probe with a concentration of 1 μM was added to the cells, and the mixture was shaken and reacted at 37° C. for 2.5 hours. After the reaction, the cells were washed three times with 500 μL of cold PBS to remove unbound probes, and then washed twice with 100 μL of 50 mM ammonium bicarbonate solution. Then surface enzyme digestion (or restriction enzyme digestion) technology is mainly used to release the reporter polypeptide from the biologic...
Embodiment 3
[0050] Example 3 Detection of HER2 protein content on breast cancer tissue sections with nucleic acid aptamer-polypeptide complex probe
[0051]36 pairs of human breast primary tumor tissues and their corresponding paracancerous tissues were cut into 6 μm tissue sections on a frozen microtome treated with 100% alcohol and 100% methanol, and 200 μL of binding buffer was dropped on the tissues On the slice, shake gently at 37°C for 1 h, and then wash the tissue slice three times with binding buffer. Then, 200 μL of binding buffer containing 200 pmol probe was added and incubated at 37° C. for 2.5 h. Finally, the tissue sections were washed three times with PBS and twice with 50 mM ammonium bicarbonate solution. Incubate the probe-loaded sections with trypsin, and collect the post-reaction solution in a centrifuge tube. After centrifugation at 20,000×g at 4°C for 10 min, 2 μg of trypsin was added to the centrifuge tube, and then reacted at 37°C for 24 hours, and 10 μL of 0.1% t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com