Real-time fluorescence detection method for detecting various pathogens and application
A fluorescence and fluorophore technology, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve the problems of increased detection difficulty, detection errors, time-consuming and labor-intensive, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
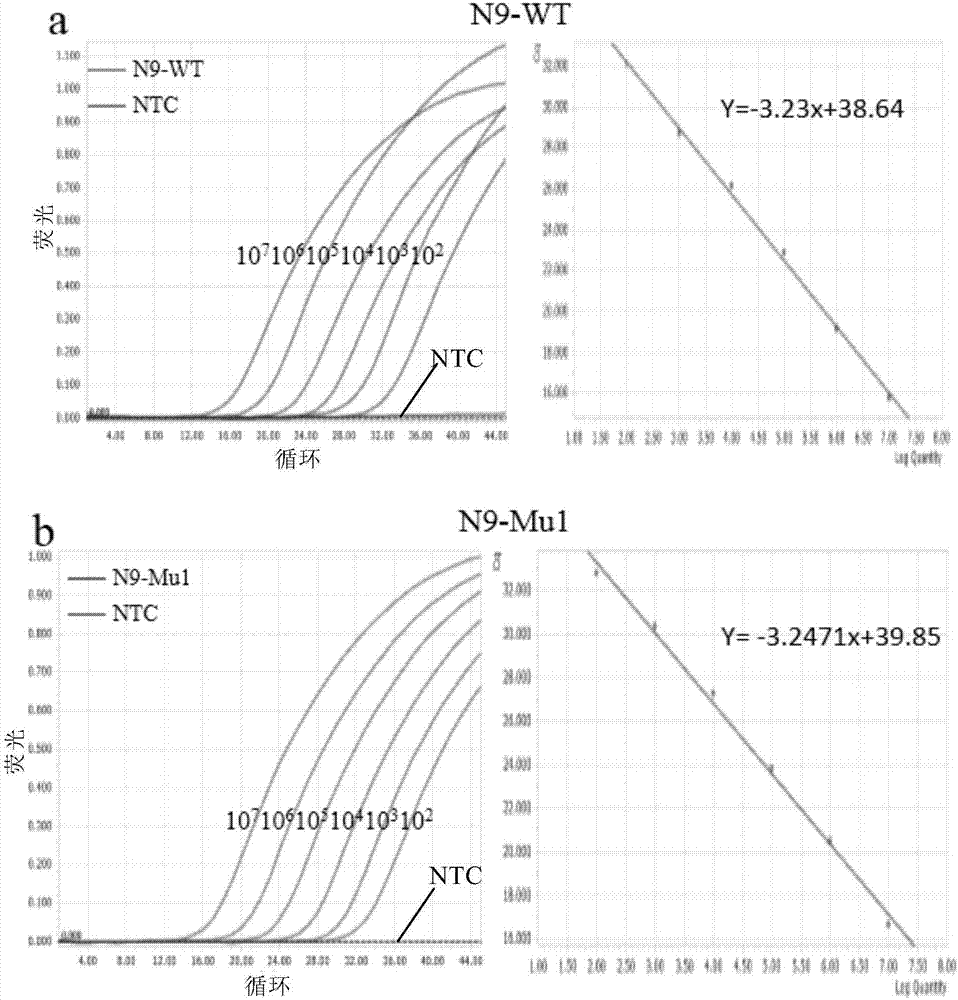
[0118] Example 1. Construction of real-time fluorescent RT-PCR detection of H7N9 influenza virus based on fluorescent primers and high-fidelity DNA polymerase system
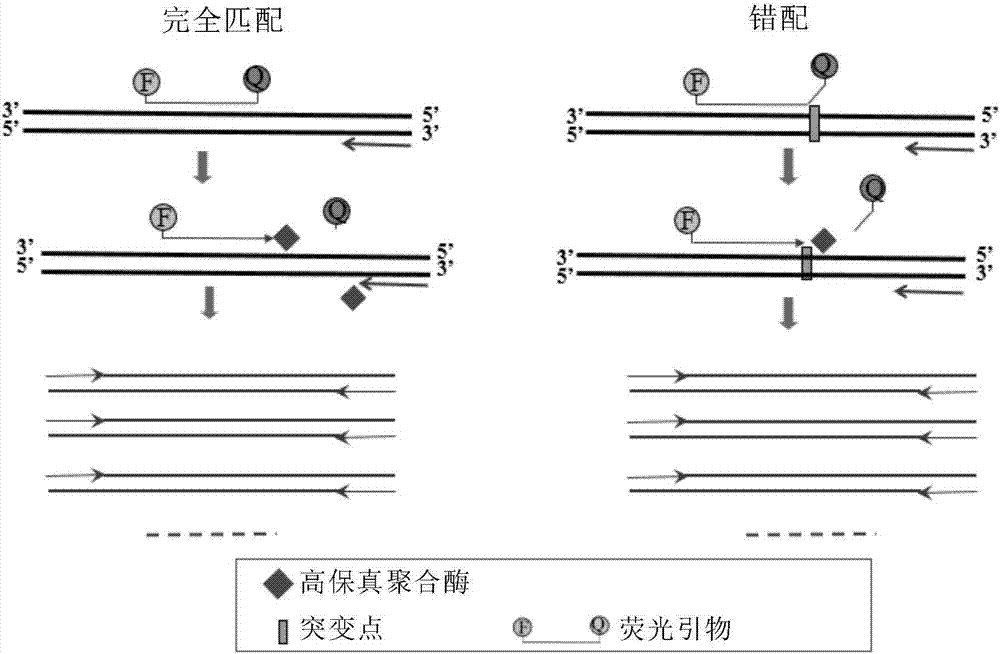
[0119] The HA fragment of influenza A virus H7N9 was selected as the RNA template, which were wild-type N9-WT and mutant N9-Mu1 sequences, respectively. There is only one single nucleotide mutation site in the two sequences, and the mutant type has only one A→G point mutation compared with the wild type. Design forward fluorescent primers and reverse ordinary primers, wherein the reverse ordinary primers are located downstream of the mutation site and are completely complementary to the wild-type and mutant templates at the corresponding positions; the forward fluorescent primers are completely complementary to the wild-type templates, and its 3' The end is located at the single nucleotide mutation site, that is, it contains a mismatch with the mutant type (as shown in Table 1). The 5' end of the forward fluore...
Embodiment 2
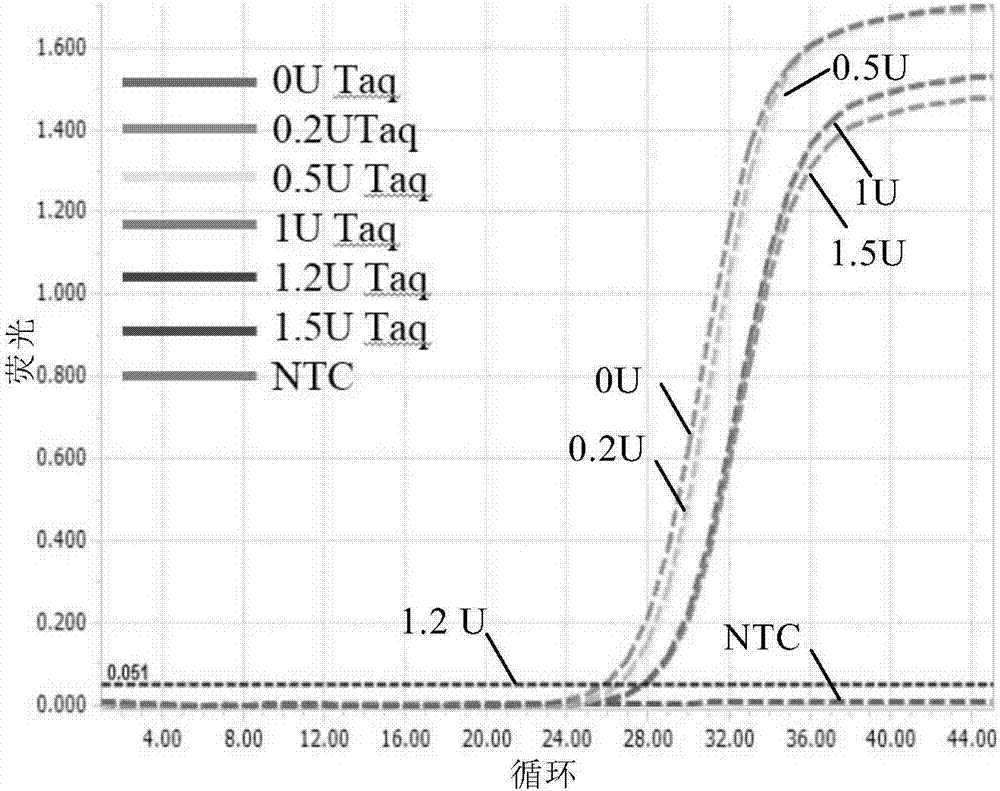
[0132] Embodiment 2, the impact of the amount of non-high-fidelity DNA polymerase on the system
[0133]Select the plasmid DNA (wild type) cloned from the HA sequence of Influenza A, and transcribe it into RNA in vitro, as a real-time fluorescent quantitative RT-PCR template, using a forward fluorescent primer exactly the same as in Example 1 And reverse general primer, add a large amount of high-fidelity enzyme to the system, add different amounts of non-high-fidelity DNA polymerase, such as 0, 0.2, 0.5, 1, 1.2, 1.5U / 25μl system, and add nuclease-free Water is the most negative control (NTC).
[0134] The reaction system is as follows
[0135]
[0136] The reaction conditions are as in Example 1.
[0137] According to the above reaction system and reaction conditions in ROCHE The reaction was carried out on a 96 real-time fluorescent quantitative PCR instrument, and the buffer solution in the reaction system and the Q5 high-fidelity enzyme were all products of NEB Comp...
Embodiment 3
[0139] Example 3, the tolerance of the detection system based on fluorescent primers and high-fidelity DNA polymerase to different mutation sites
[0140] The present invention selects human gene β-Actin (ACTB) for PCR amplification and in vitro transcription to obtain RNA standard (wild-type ACTB-WT), and designs a mutant template with only a single nucleotide mutation (A→G) sequence (ACTB-Mu1), mutant template sequence (ACTB-Mu2) with only a single nucleotide mutation (A→U), mutant template sequence with a single nucleotide mutation (A→C) (ACTB- Mu3). Forward fluorescent primers and reverse ordinary primers were designed respectively, wherein the reverse ordinary primers were located downstream of the mutation site and completely matched with the wild-type and mutant templates at the corresponding positions; the 3' ends of the four forward fluorescent primers were located at the point mutations ( As shown in table 2). The 5' end of the forward fluorescent primer was modifi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com