DNA barcode identification method of medical and edible material spina date seed and its fake product
A technology of jujube seed and barcode, which is applied in recombinant DNA technology, DNA/RNA fragment, and microbial determination/inspection.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0069] Example 1 Intraspecific Variation Analysis of Suanzaoren Medicinal Material
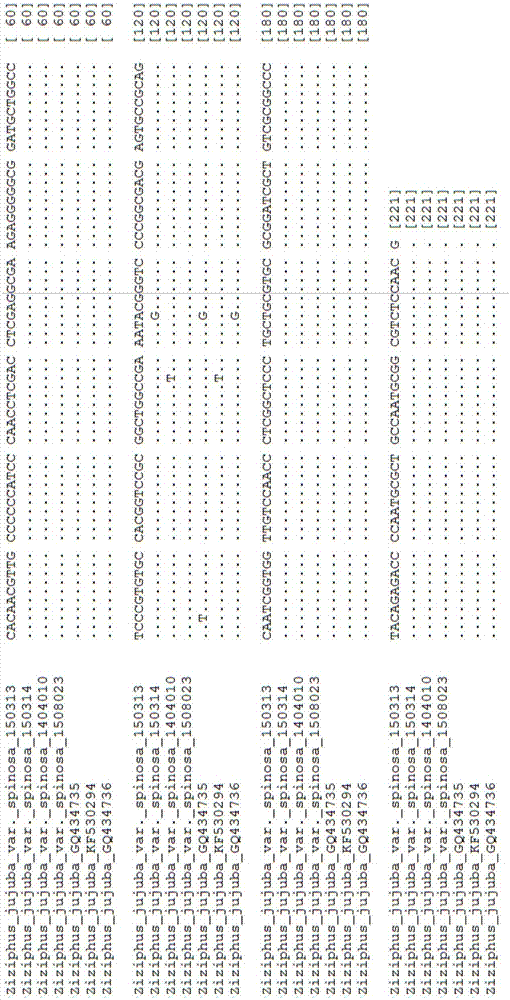
[0070] After the comparison of 20 sequences from different sources of Zianzioren, the length of the psbA-trnH sequence was 321bp, and the GC content was 26.3%. There are 8 haplotypes at positions 232, 246, 258, 261, 268, 274, and 281. 5 samples of haplotype H1 and H2, same as 06AZ820151126363 and 06AZ820151126366 respectively; 4 samples of H3 haplotype, same as 06AZ820151126375; 2 samples of H4 haplotype, same as 06AZ820151126372; H5, H6, H7, H8 haplotype samples Both are 1, respectively 06AZ820151112501, EU075108, 06AZ820151126365 and 06AZ820151126367 (Table 2). The genetic distance was calculated based on the K2P model. The average K2P distance within the species of the psbA-trnH sequence of Suanzaoren medicinal material was 0.003, and the maximum K2P genetic distance within the species was 0.017.
[0071] Table 2 Intraspecific variation sites of psbA-trnH sequence in Suanzaoren
[0072] ...
Embodiment 2
[0074] Example 2 Interspecies Variation Analysis of Suanzaoren Medicinal Material and Its Counterfeit Products
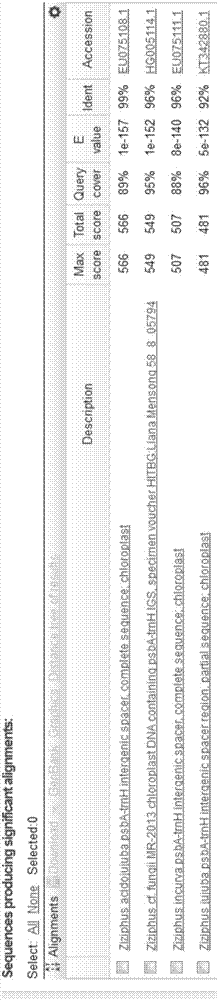
[0075] After alignment of 18 sequences from samples from different sources in C. yunnanensis, the length of psbA-trnH sequence was 349bp, and the GC content was 25.4%. The minimum inter-species K2P distance of the psbA-trnH sequences of Suanzaoren and its adulterant, C. dianthus, was 0.310, and the average inter-species K2P distance was 0.321. The minimum interspecific distance between Suanzaoren and its counterfeit products was between Suanzaoren and Hovenia dulcis, which was 0.091, and the average K2P genetic distance was 0.093. The minimum interspecific K2P genetic distances of the psbA-trnH sequences of Suanzaoren and its counterfeit products were greater than the maximum intraspecific K2P genetic distances (Table 3).
[0076] Table 3 Intraspecific genetic distance of Jujube jujuba and its interspecific genetic distance with counterfeit varieties
[0077]
Embodiment 3
[0078] Example 3 Systematic cluster analysis of Jujube Seed Medicinal Materials and its counterfeit products
[0079] Based on the psbA-trnH sequence, the NJ phylogenetic clustering tree of jujube seed and its counterfeit products was constructed (see Figure 5 ). In the picture, jujube seed and its counterfeit products are clustered into different branches, which can be clearly distinguished from the counterfeit products. C. yunnanensis, Hovenia dulcis, Lentil and Bauhinia all clustered into one branch, showing good monophyletic.
[0080] In the present invention, a total of 5 species including Jujube Seed and its common counterfeit products, Lizaoren, Hovenia dulcis, Lentil and Bauhinia are selected for DNA barcode research. The intraspecific and interspecific genetic distance analysis of Suanzaoren and its counterfeit products showed that the maximum intraspecific genetic distance of 0.017 was much smaller than the minimum interspecific distance of 0.091 between Suanzaore...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com