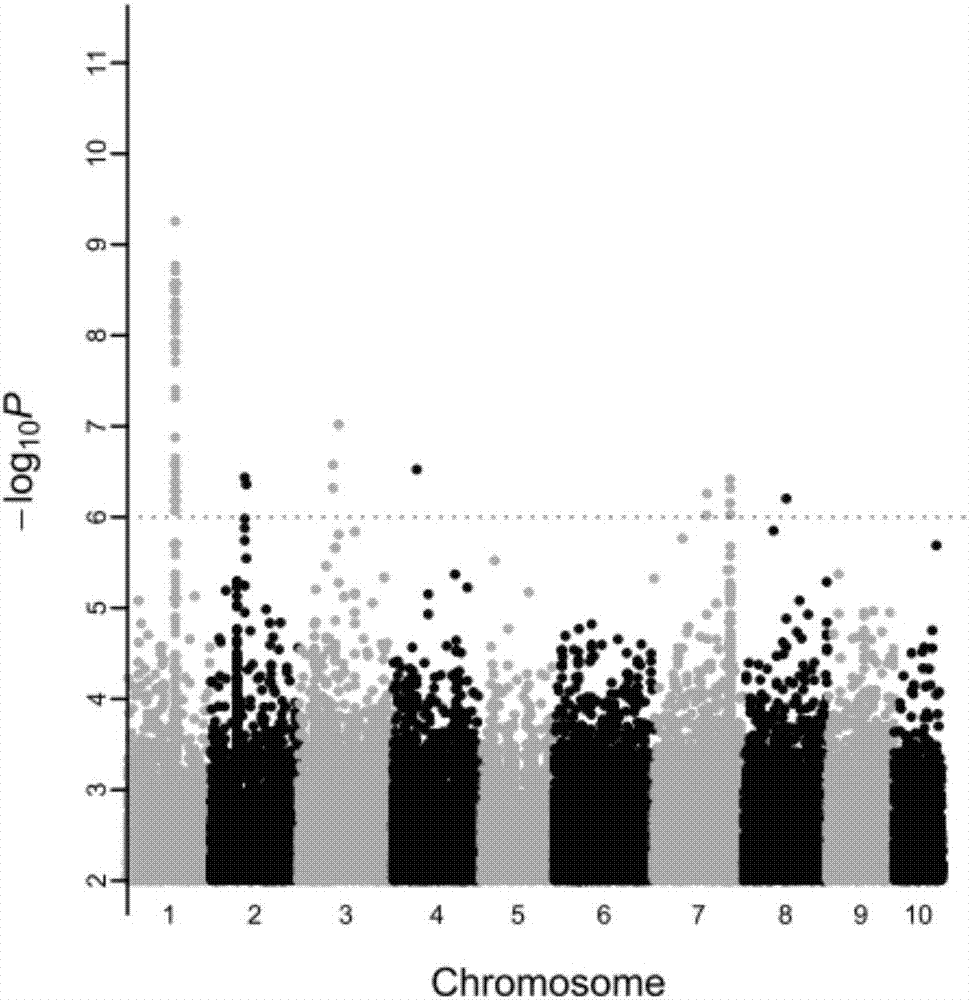
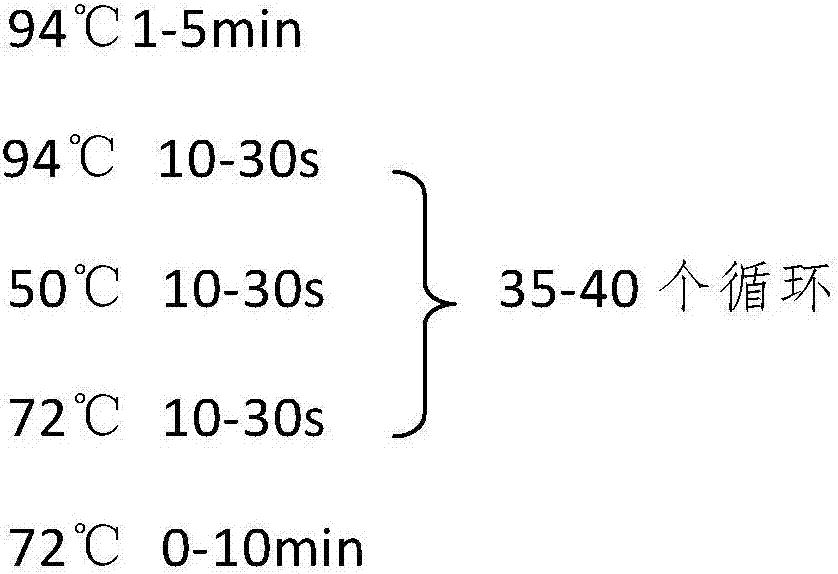
Method for screening pacific oysters capable of increasing content of unsaturated fatty acid C20:3omega6 and related SNP (Single Nucleotide Polymorphism) primer pair
A technology of unsaturated fatty acids and long oysters, applied in the field of genetic engineering and genetic breeding, can solve the problems of inability to understand the genetic regulation mechanism and the small number of markers
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] a) Collection of samples: Collect 100 individuals from wild populations in Jiaonan where seedlings were hatched at the same time, dissect them, take adductor muscle and remaining tissues, and store them at -80°C after quick-freezing with liquid nitrogen.
[0041] b) Extraction of DNA: extract the genomic DNA of 100 samples and measure the concentration by ultraviolet absorbance photometry, and dilute the genomic DNA to 10-20ng / uL with sterile water according to the measured concentration;
[0042] c) Using SnaPshot to detect the genotype of the SNP site: (1) Using specific primers to perform peripheral amplification. Take the above long oyster genomic DNA as a template, and use primers F and R to prepare a reaction system: 1uL of genomic DNA, 5uL of general PCR mix, 0.2uL of primers F and R, 3.6uL of sterilized double-distilled water; the reaction system can be scaled up year-on-year; The primer sequences are:
[0043] Upstream primer F: 5'-ACGTCACAAGTAGGTGTTTAGAGA-3' ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com