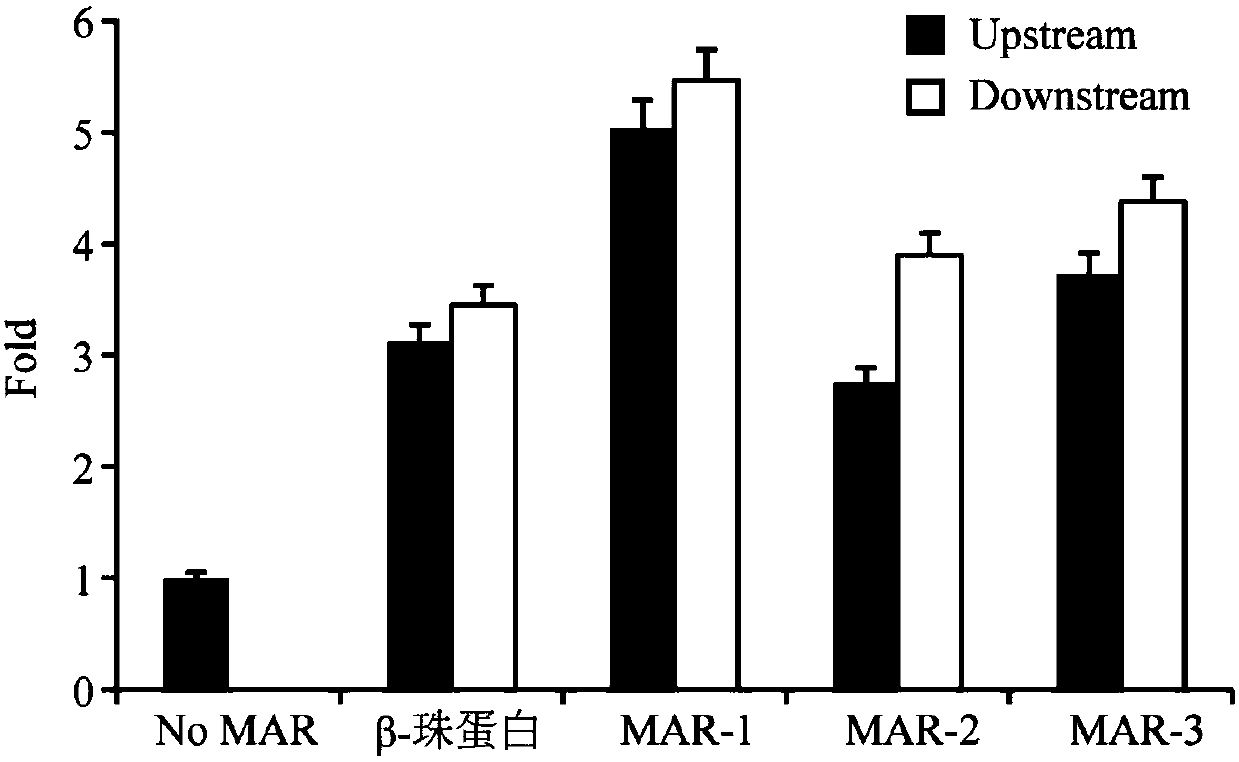
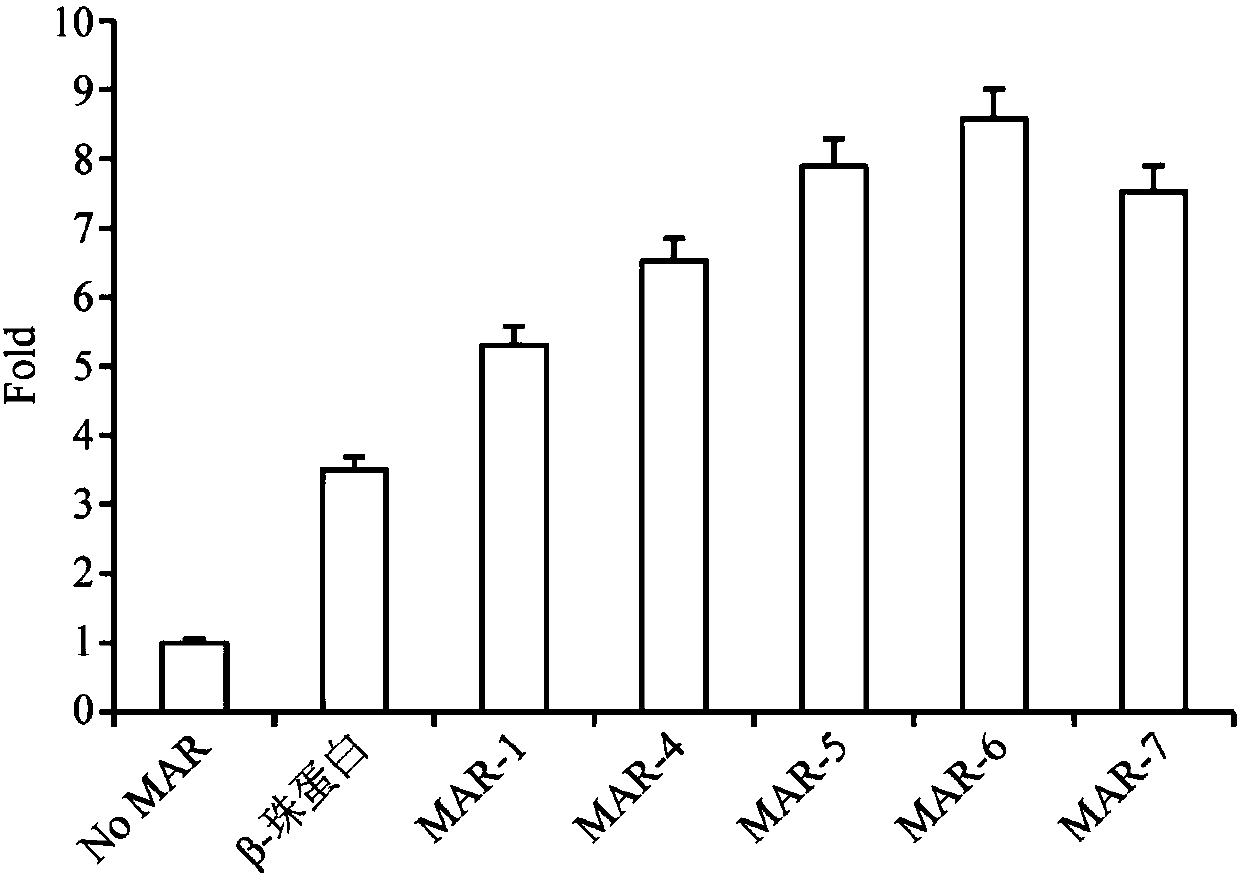
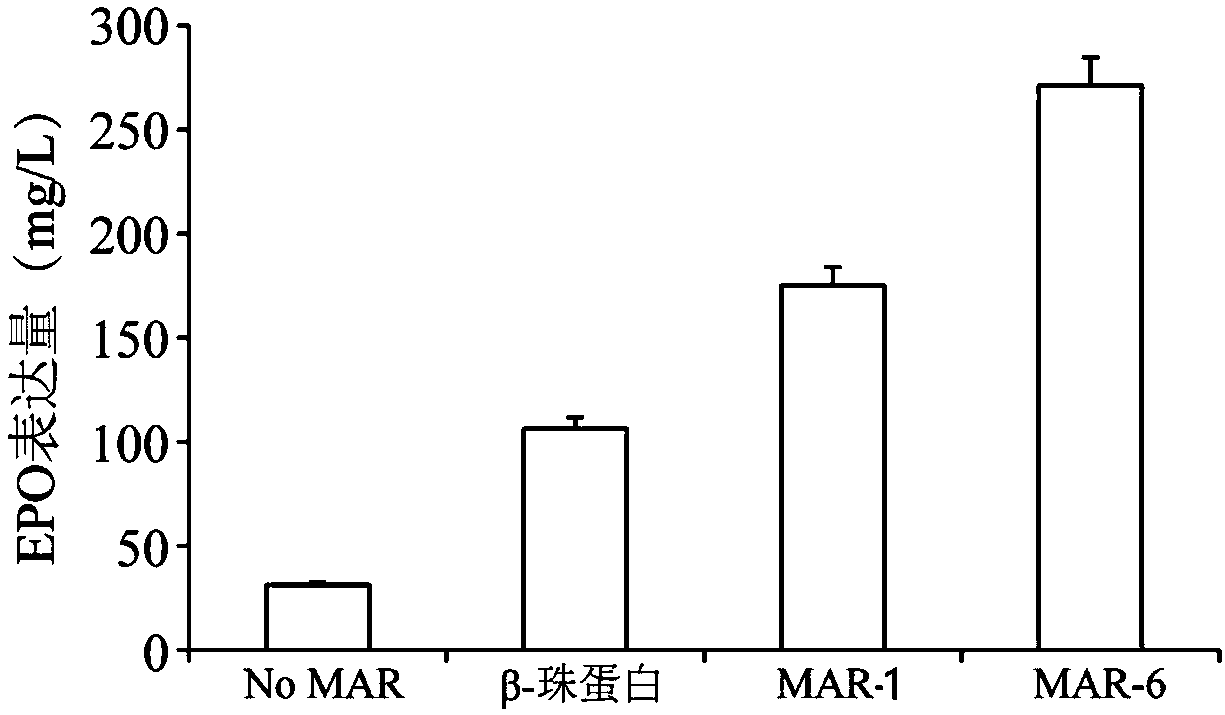
Artificially synthesized MAR (Matrix Attachment Region) fragment, expression vector, expression system and application thereof
A technology for artificially synthesizing and expressing vectors, applied in the directions of DNA/RNA fragments, vectors, and the introduction of foreign genetic material using vectors, which can solve the problems of increasing the difficulty of vector construction and reducing the efficiency of plasmid transfection, and achieve stable and long-term expression, reducing The effect of expressing differences and improving stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] 1) Design and synthesis of artificial MAR fragments.
[0028] The MAR sequence is an AT-rich DNA sequence. Although it has no consensus sequence, the MAR sequence has typical characteristic motifs such as A-box, T-box, ARS, Unwinding sequence, Curved DNA, Oligo A-T tracts, A- tracts, T-tracts, Stem loops, Curved DNA, SATB1, ATF site, Topoisomerase II, CEBP, FAST, Hox, NMP4, GSH, etc. (as shown in Table 1).
[0029] Table 1 Characteristic motifs of MAR
[0030]
[0031] In Table 1, Y is T / C, W is A / T, R is G / A, N is A / T / G / C, K is G / T, S is G / C, and M is A / C.
[0032]According to literature reports (Genome-wide prediction of matrix attachment regions that increase gene expression in mammalian cells. Nat. Methods. 2007; Positional effects of the matrix attachment region on transgene expression in stably transfected CHO cells. Cell Biol. Int. 2010);
[0033] (Molecular characterization of a human matrix attachment region that improves transgene expression in CHO cells....
Embodiment 2
[0036] An expression vector containing synthetic MAR-1 upstream of the CMV promoter was constructed.
[0037] For the convenience of cloning, NruI (TCGCGA) and MluI (ACGCGT) restriction sites were respectively inserted at the 5' and 3'-ends of the synthesized MAR-1 fragment.
[0038] The synthesized MAR-1 sequence was digested with NruI / MluI, and the pIRES-Neo plasmid DNA vector (purchased from Clontech Biological Company) was simultaneously digested with NruI / MluI. The digestion results were identified by agarose gel electrophoresis, and the digested MAR sequence fragment and pIRES-Neo linear plasmid DNA were recovered from the gel.
[0039] The double enzyme digestion system of MAR-1 sequence is: MAR-1 sequence 10μL (1μg / μL), 10×NE Buffer 3.1 3μL, NruI / MluI (10U / μL) each 1.0μL, make up water to 30μL; enzyme digestion conditions are : 37°C, enzyme digestion for 3min.
[0040] The double enzyme digestion system of pIRES-Neo plasmid is: pIRES-Neo plasmid 5 μL (1 μg / μL), 10×NE...
Embodiment 3
[0043] The construction method is the same as that in Example 2, except that the fragment is replaced by the MAR-2 fragment (sequence shown in SEQ ID NO.2), and the correctly constructed plasmid is named pIRES-MAR1-2.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com