Recombinant contagious ecthyma oncolytic virus and preparation method and application thereof
An oncolytic virus, infectious technology, applied in the field of genetic engineering, to achieve the effect of shortening the screening time and easy operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
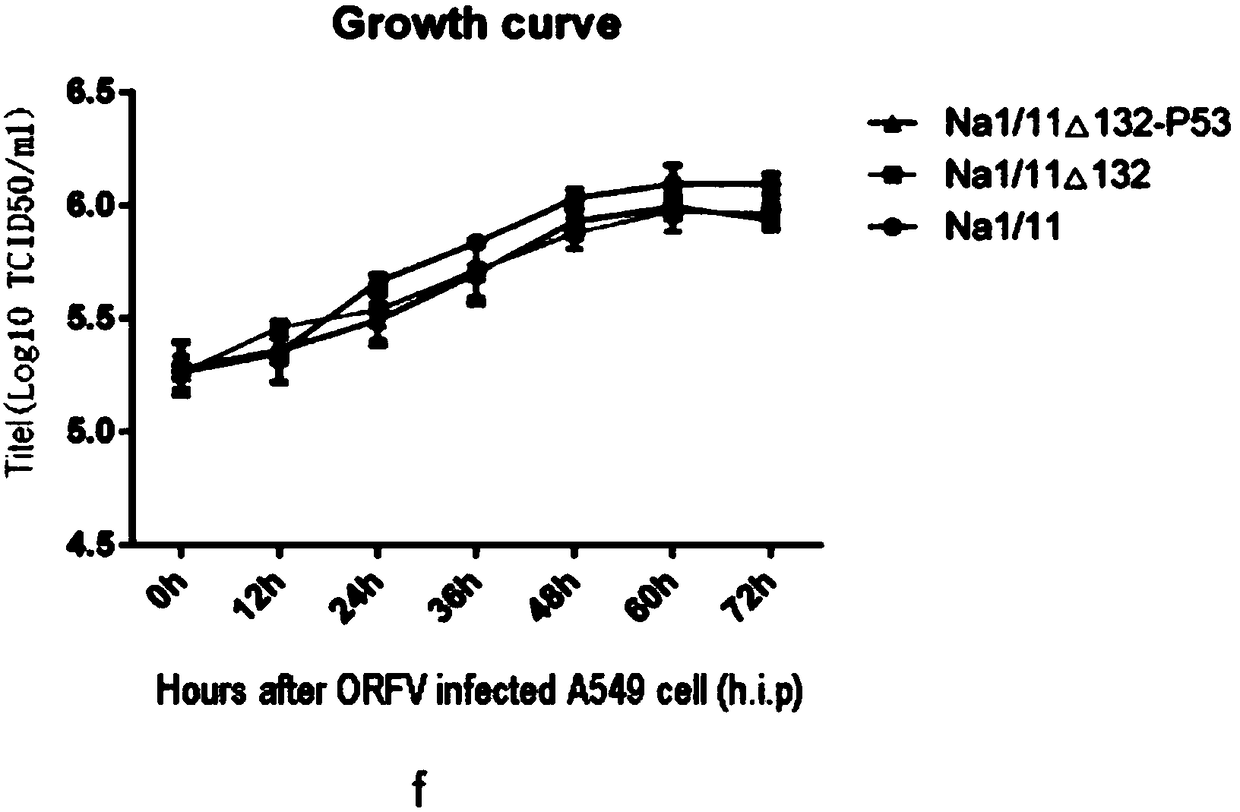
Image
Examples
preparation example Construction
[0064] The preparation method of the recombinant infectious pustular oncolytic virus of the present invention comprises the following steps:
[0065] (1) overlap PCR amplifies the gene p53 of the protein described in claim 7;
[0066] (2) Connect to the pSPV-EGFP vector, insert the p53 gene into a recombinant plasmid to obtain the recombinant plasmid p53-pSPV-EGFP, and then insert the left and right sides of the ORFV132 gene to construct the shuttle plasmid ORFV132LF-p53-pSPV-EGFP-ORFV132RF; Recombinant technology inserts p53 and EGFP genes into position 132 of the viral genome, thereby knocking out the ORFV132 gene; PCR and double digestion of ORFV132LF-p53-pSPV-EGFP-ORFV132RF: the recombinant plasmid band is at 6500bp, PCR amplification The band is at 700bp, and the two bands are 5600bp and 700bp respectively after digestion; the predicted ORFV132LF-p53-pSPV-EGFP-ORFV132RF is 6392bp, and ORFV132RF is 676bp, and the two bands after double digestion are 5649bp and 5649bp respe...
Embodiment 1
[0081] Construction of recombinant plasmid p53-pSPV-EGFP
[0082] (1) Amplification and identification of the target gene p53
[0083] According to the requirements of the seamless cloning kit, using p53 as a template, use the Primer Premier5 software to design PCR primers (the underlined part is the restriction site):
[0084] Hp53-Fw:
[0085] 5'-CAGTGACGCCTCGAG GAATTC ATGGAGGAGCCGCAGTCAGA-3' (EcoRI); SEQ ID No.1
[0086] Hp53-Rv:
[0087] 5'-GCTCACCATGGTGGC GAATTC GTCTGAGTCAGGCCCTTCTGTCTT-3' (EcoRI). SEQ ID No. 2
[0088] Perform PCR reaction (50 μL) according to the following system as shown in Table 1:
[0089] Table 1
[0090]
[0091] Reaction conditions:
[0092]
[0093] The PCR products were identified by agarose electrophoresis and recovered by tapping the gel.
[0094] (2) pSPV-EGFP digestion and rubber tapping recovery are shown in Table 2:
[0095] Table 2
[0096]
[0097] Digest at 37°C for 2 hours according to the above system, and recover...
Embodiment 2
[0107] Construction of recombinant plasmid ORFV132LF-p53-pSPV-EGFP
[0108] The method for constructing the recombinant plasmid ORFV132LF-p53-pSPV-EGFP is the same as 1. The upstream and downstream primers are respectively (the underlined part is the restriction site):
[0109] NA1 / 11-ORFV132L-Fw: 5'-AGTAGGCCTGCGCGCAAGCTTCGTCTTCTCCCGCTGGATAAA-3'(HindⅢ) SEQ ID No.3
[0110] NA1 / 11-ORFV132L-Rv: 5'-GACCTGCAGGCATGCAAGCTTGCCTCACCCTTAAAAGTTGG-3'(HindⅢ) SEQ ID No.4;
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com