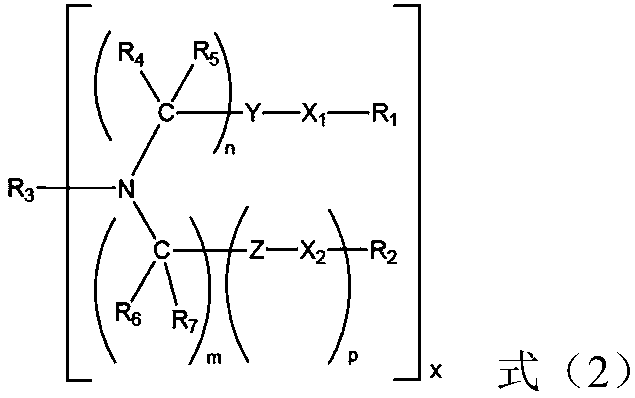
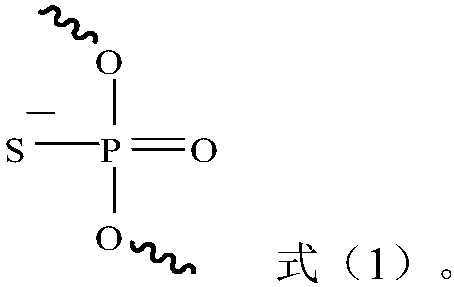Small interfering nucleic acid and pharmaceutical composition and application thereof
A nucleotide and nucleotide sequence technology, applied in the field of biomedicine, can solve the problems of poor stability of siRNA, easy to be degraded by nucleases, differences between target nucleic acid species, etc., to achieve inhibitory content, prevention and/or treatment of blood lipids abnormal effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example 1
[0106] The sequence of siRNA is shown in Table 2, and the nucleotide sequence of the sense strand numbered siAP-1 is shown in SEQ ID NO.6, where the nucleotide sequence of positions 1-19 is similar to the human APOC3 mRNA sequence (NM_000040.1). The target nucleic acid shown in SEQ ID NO.1 is the same; the nucleotide sequence of the antisense strand of the siRNA is shown in SEQ ID NO.7, and the nucleotide sequence of positions 1-19 is the same as that shown in SEQ ID NO.1 in Table 1. The indicated target nucleic acids are complementary. The nucleotide sequence of the sense strand numbered siAP-2 is shown in SEQ ID NO. 8, wherein the nucleotide sequence of positions 1-19 is the same as the target nucleic acid shown in SEQ ID NO. 2 in the APOC3 mRNA sequence; The nucleotide sequence of the antisense strand is shown in SEQ ID NO. 9, wherein the nucleotide sequence at positions 1-19 is complementary to the target nucleic acid shown in SEQ ID NO. 2 in Table 1. The nucleotide sequen...
Embodiment 1
[0114] This example is used to detect the inhibition rate of the siRNA obtained in Preparation Example 1 on the expression level of APOC3 mRNA in vitro.
[0115] The human hepatoma cell line Huh7 was inoculated into a 24-well plate with DMEM complete medium containing 10% fetal bovine serum at a density of 4×10 5 Cells / well, 0.5mL medium per well, cultured overnight at 37°C.
[0116] Aspirate the cell culture solution in the 24-well plate, and add 0.5 mL of Opti-MEM serum-free medium to each well. Dilute 1.5 μL of the siRNA in preparation example 1 at a concentration of 20 μM with 50 μL of Opti-MEM serum-free medium; add 1 μL of Lipofectamine TM 2000 (Invitrogen) diluted in 50μL Opti-MEM serum-free medium, mixed and incubated at room temperature for 5 minutes; mixed diluted siRNA and diluted Lipofectamine TM 2000, mix gently and let stand at room temperature for 20 minutes to allow complex formation. The final mixed solution was added to a 24-well plate seeded with Huh7 cells ...
preparation example 2
[0125] The siRNAs obtained after chemical modification of the siRNA sense strand and antisense strand numbered siNC are shown in Table 5, numbered as siNC-M; 3 groups of siRNAs obtained after chemical modification of the siRNA sense strand and antisense strand numbered siAP-1 As shown in Table 5, they are numbered siAP-1-M1, siAP-1-M2, and siAP-1-M3. The three groups of siRNAs obtained after chemical modification of the siAP-2 siRNA sense strand and antisense strand are shown in Table 5, respectively numbered siAP-2-M1, siAP-2-M2, and siAP-2-M3. The three groups of siRNAs obtained after chemical modification of the siAP-3 siRNA sense strand and antisense strand are shown in Table 5, respectively numbered siAP-3-M1, siAP-3-M2, and siAP-3-M3. The three groups of siRNAs obtained after chemical modification of the siAP-4 siRNA sense strand and antisense strand are shown in Table 5, respectively numbered siAP-4-M1, siAP-4-M2, siAP-4-M3.
[0126] Where m represents the pentose group o...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
| particle diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com