Method for detecting NOD1 gene single nucleotide polymorphism with HRM (High Resolution Melting) technology
A single nucleotide polymorphism and technology detection technology, applied in the field of HRM technology detection of NOD1 gene single nucleotide polymorphism, can solve the problems of high reaction conditions, complicated operation process, cumbersome test process, etc., and achieve high sensitivity And the effect of high specificity, good repeatability, and shortened detection time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] The method for detecting the SNP site rs2075820 of the NOD1 gene in the subject by using the high-resolution melting curve (HRM) method includes obtaining sample DNA and providing reagents required for sequencing. Refer to Example 2 for specific operation steps. The reagents include: Whole Blood Genomic DNA Extraction Kit (QIAGEN), HRM Detection Kit FP210 (Tiangen Biochemical Technology Co., Ltd., Beijing, China), primers for detecting rs2075820 site, standard products and negative control products. Among them, the primers for detecting the rs2075820 site are:
[0025] NOD1-F-HRM:GCTTCAAGGAAAGTGACAGGC;
[0026] NOD1-R-HRM: CAGGTCCAAGTCCGAGTGC.
[0027] Standards are GG genotype, GA genotype and AA genotype DNA samples; negative control: DNA-free solution.
[0028] Detection system PCR reaction solution includes:
[0029]
[0030] The PCR reaction procedure is:
[0031]
[0032] The present invention also adopts the sequencing method to further verify the resul...
Embodiment 2
[0042] Operation procedure of blood / cell / tissue genomic DNA extraction kit (QIAGEN):
[0043] (1) Genomic DNA extraction from whole blood
[0044] 1) Add 20μl QIAGEN Protease (or proteinase K) to the bottom of a 1.5ml centrifuge tube. .
[0045] 2) Add 200 μl of plasma to the centrifuge tube.
[0046] 3) Add 200 μl Buffer AL and shake for 15 seconds. (Note: Do not add QIAGEN Protease or proteinase K directly to Buffer AL. If the sample size is large, increase QIAGEN Protease and Buffer AL proportionally.)
[0047] 4) Water bath at 56°C for 10 minutes, and then briefly centrifuge to remove the liquid on the inner edge of the centrifuge tube cover.
[0048] 5) Add 200 μl of ethanol (96%-100%), shake for 15 s, and briefly centrifuge to remove the liquid along the inner edge of the centrifuge tube cover.
[0049] 6) Carefully add the mixture (including the precipitate) obtained above to the QIAamp Mini spin column (without wetting the rim). Put the spin column into a 2ml coll...
Embodiment 3
[0096] Embodiment 3 clinical sample detection
[0097] Forty clinical samples to be tested were taken, and the genome was extracted, reagents were prepared and tested according to the methods and reagents described in Examples 1 and 2.
[0098] Add 1 μL of each sample to the detection system PCR reaction solution. At the same time, make a copy of each standard product. Detect with a fluorescent PCR instrument, and the time is 120 minutes.
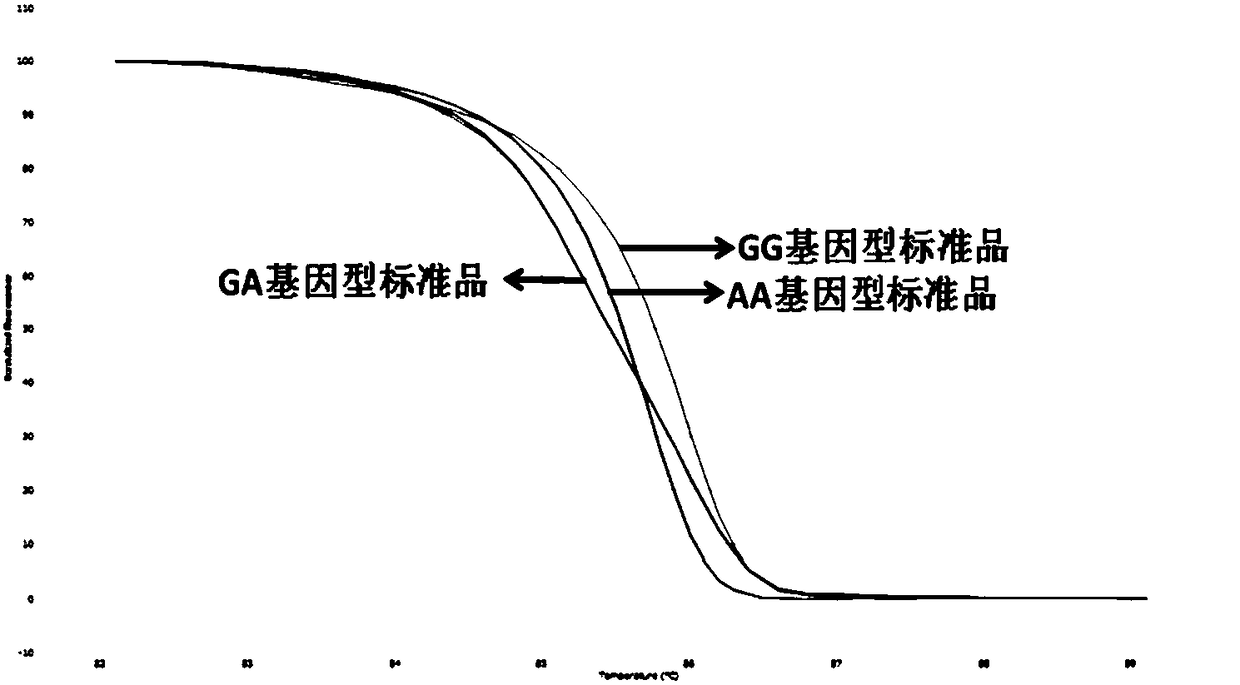
[0099] Such as figure 1 Shown are the HRM results for Standard AA, Standard GA, and Standard GG.
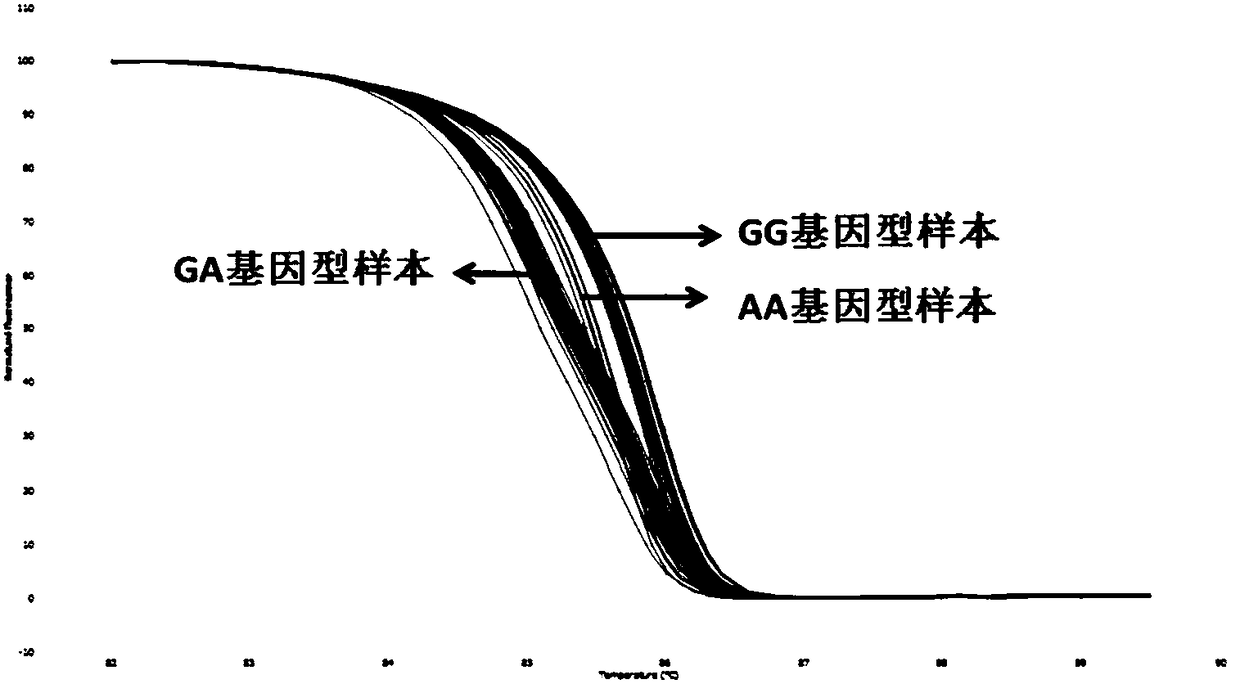
[0100] Such as figure 2 Shown are the HRM results of 40 clinical samples and standards.
[0101] According to the HRM results, the genotypes at the rs2075820 locus of the NOD1 gene of 40 screening samples were interpreted as shown in Table 1 below.
[0102] Table 1
[0103]
[0104]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com