Oidium heveae Steinmann in-vivo promoter WY51 and application thereof
A technology of rubber tree powdery mildew and endogenous promoter is applied in the field of genetic engineering and can solve problems such as strengthening
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Embodiment 1, PCR amplification of WY51 promoter fragment
[0035] Use the fungal genomic DNA extraction kit (OMEGA, D3390-01) to extract the genomic DNA of Powdery mildew Hevea (provided by the Key Laboratory of Sustainable Utilization of Tropical Biological Resources in Hainan Province), and design a pair of specific amplification according to the sequence of the WY51 promoter Primers (upstream primer WY51F, plus restriction site HindⅢ and protective bases, downstream primer WY51R, plus restriction site BamHI and protective bases). Using the extracted genomic DNA of Erysiphaea hevea as a template, high-fidelity Ex Taq polymerase (TRANSGEN, AP122) was used for PCR amplification. As shown in Table 1.
[0036] Table 1 PCR system for gene promoter amplification
[0037]
[0038] The PCR amplification program was as follows: pre-denaturation at 94°C for 5 min, then denaturation at 94°C for 60 s, annealing at 55°C for 50 s, extension at 72°C for 60 s, 35 reaction cycle...
Embodiment 2
[0041] Embodiment two, the construction of pGEM-T easy-WY51 recombinant vector
[0042] The PCR amplification product obtained above was transformed into Escherichia coli (TRANSGEN, CD201) by T / A cloning (pGEM-T easy plasmid, PROMEGA, A1360), and positive clones were picked and sequenced, which proved to be accurate.
[0043] Among them, the connection conditions of T / A clone are as follows:
[0044]T / A connection system: 10ul
[0045] pGEM-T Easy Vector (PROMEGA, A137A): 1ul
[0046] 2×Rapid ligation Buffer: 5ul
[0047] PCR amplification product (recovered insert): 2ul
[0048] T4DNA ligase: 1ul
[0049] wxya 2 O: 1ul
[0050] First place at room temperature for 1 hour, then ligate overnight at 4°C to obtain pGEM-T easy-WY51 recombinant vector. The product after the above connection was transformed into Escherichia coli as follows:
[0051] Take out 100 μl of DH5α (Transgene, CD201) competent cells prepared according to the calcium chloride method shown in "Molecular...
Embodiment 3
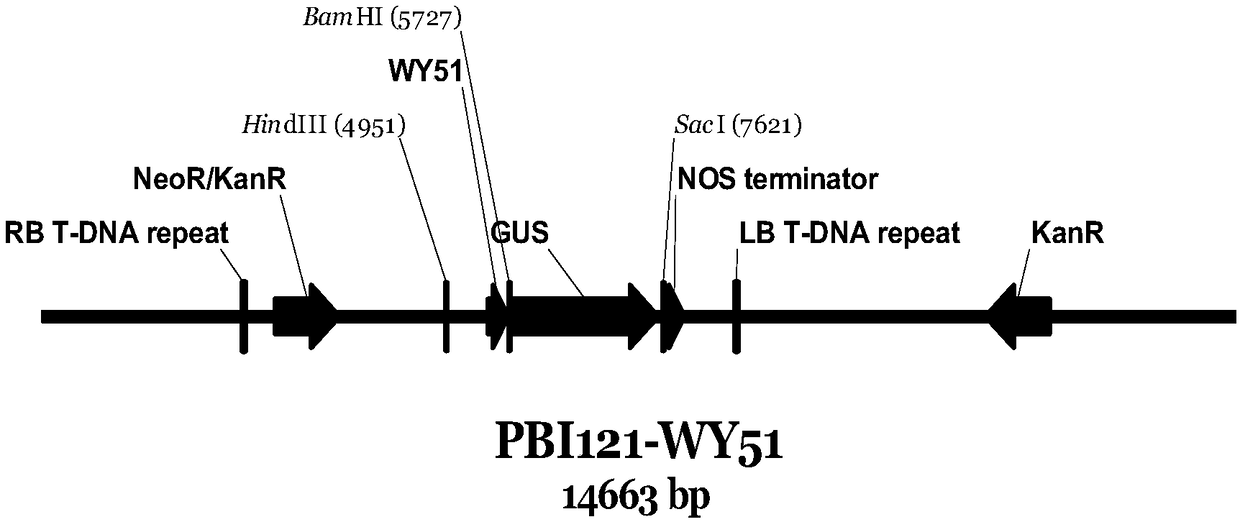
[0053] Example 3, Construction of PBI121-WY51 recombinant vector
[0054] Pick a single colony of the DH5α-WY51 strain obtained from the above construction and shake the bacteria overnight at 37°C at 220 rpm. The plasmid is extracted with the OMEGA plasmid mini-extraction kit (D6943-01), and then HindⅢ (NEB, R0104S) and BamHI ( NEB, R0136V) restriction endonuclease was used for double digestion, and the digested product was recovered with the OMEGA recovery kit (D2500-01) to recover the WY51 promoter fragment.
[0055] The recovered product obtained above was ligated with the PBI121 plasmid (TIANNZ, 60908-750y), and then transformed into Escherichia coli, and positive clones were picked and sequenced, which proved to be accurate.
[0056] Among them, the connection conditions of T / A clone are as follows:
[0057] T / A connection system: 10ul
[0058] PBI121Vector: 1ul
[0059] 10×T4DNA Ligase Buffer: 1ul
[0060] Recovered product (WY51 promoter fragment): 6ul
[0061] T4D...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com