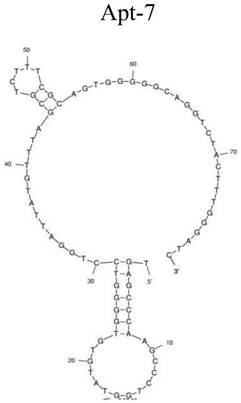
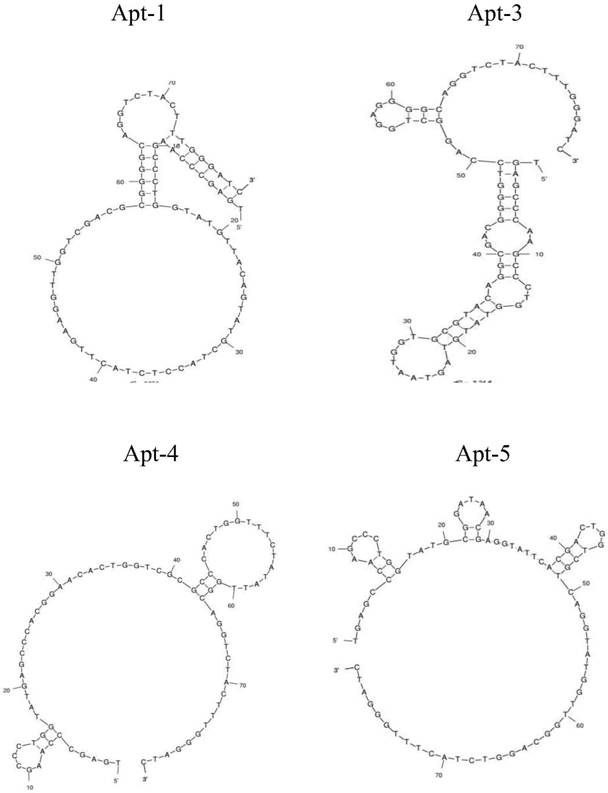
A group of oligonucleotide aptamers that specifically recognize Escherichia coli O157:H7 in different growth stages
An oligonucleotide and Escherichia coli technology, which is applied in the field of food safety biology, can solve the problems such as the inability to meet the detection requirements, and achieve the effects of easy labeling, stable properties and convenient synthesis.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Example 1: Screening of aptamers
[0026] 1. Synthesize random single-stranded DNA library and primers (completed by American Industrial Integrated DNA Technologies)
[0027] Random ssDNA library:
[0028] 5’-TGA GCC CAA GCC CTG GTA TG-N40-GGC AGG TCT ACT TTG GGA TC-3’
[0029] 5'-carboxyfluorescein-labeled upstream primer: 5'-FAM-TGA GCC CAA GCC CTG GTA TG-3'
[0030] 5' phosphorylation downstream primer: 5'-P-GAT CCC AAA GTA GAC CTG CC-3'
[0031] The random ssDNA library and primers were prepared with TE buffer as a 100uM stock solution and stored at -20°C for future use.
[0032] 2. Selection and processing of bacteria
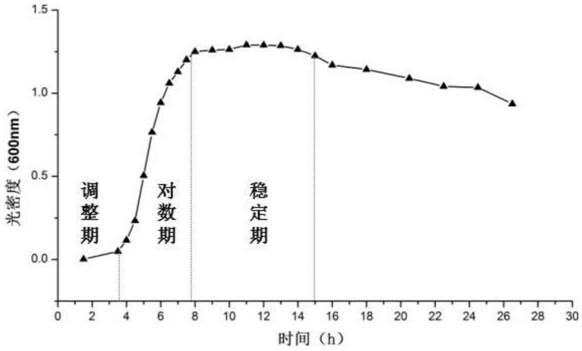
[0033] (1) Determination of the growth curve of Escherichia coli O157:H7
[0034] Escherichia coli O157:H7 was cultured in LB (tryptone 10g / L, yeast extract 5g / L, sodium chloride 10g / L, pH7.4) liquid medium, 37°C, 120r / min shaker culture, The OD value of the bacterial solution was measured at regular intervals. According to the value of each ...
Embodiment 2
[0047] Example 2: Aptamer affinity and specificity testing
[0048] (1) Determination of dissociation constant
[0049] The synthesized 9 aptamers were diluted with TE buffer to prepare a 10 pmol / L solution, and stored at -20°C for later use. The affinity of 9 aptamers was analyzed by BDFACS Calibur flow cytometer. Different volumes of 10 pmol / L aptamers were added to 500 μL of BB binding buffer and diluted to different concentration gradients (10, 50, 100, 150, 200 nmol / L), denatured at 95 °C for 10 min, and immediately cooled at 0 °C for 10 min. The aptamer solution was added to the treated E. coli O157:H7 and incubated at 37°C with slow shaking for 1 h.
[0050] It was then washed with BB buffer and resuspended in 500 μL BB buffer for flow cytometry analysis. In the flow cytometry analysis, the fluorescence intensity of the blank sample (without aptamer) was adjusted first, and then the forward scatter, side scatter and fluorescence intensity of the sample were measured ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com