Early stage molecular detection and identification method and application for eight kinds of oophagous trichogrammae in trichogrammatidae family
A technology of molecular detection and identification method, which is applied in the field of species identification, can solve problems such as time-consuming, and achieve the effects of fast detection, wide application range, and simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
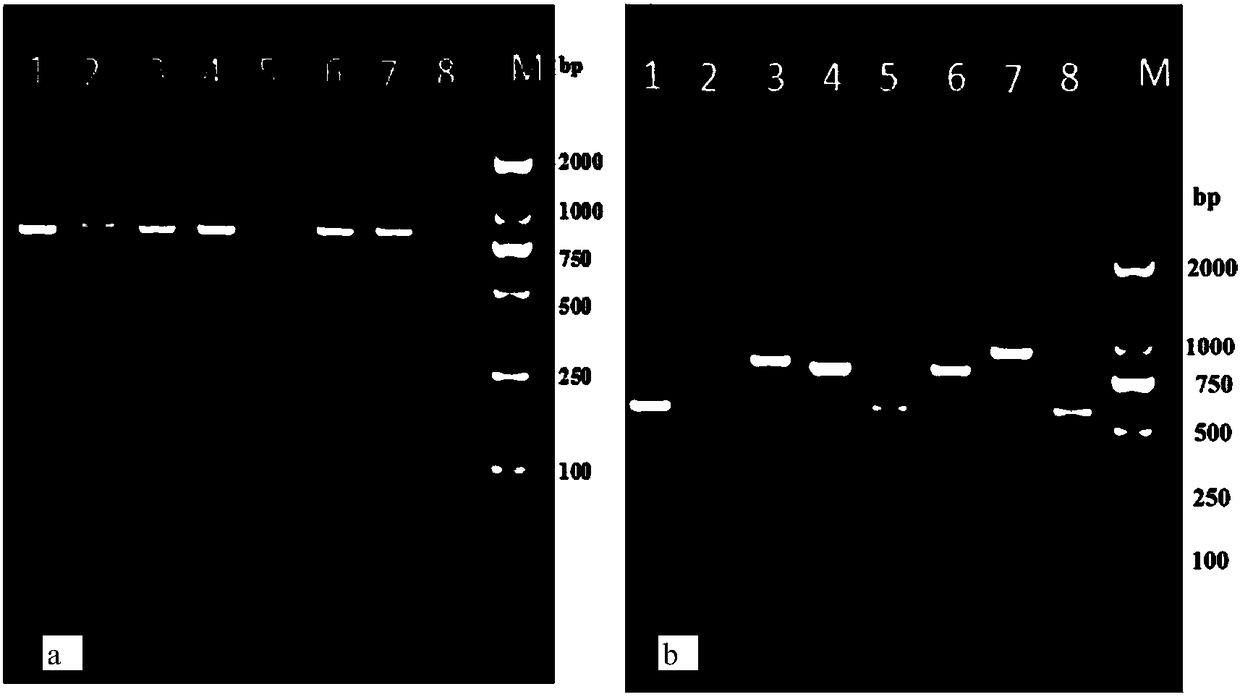
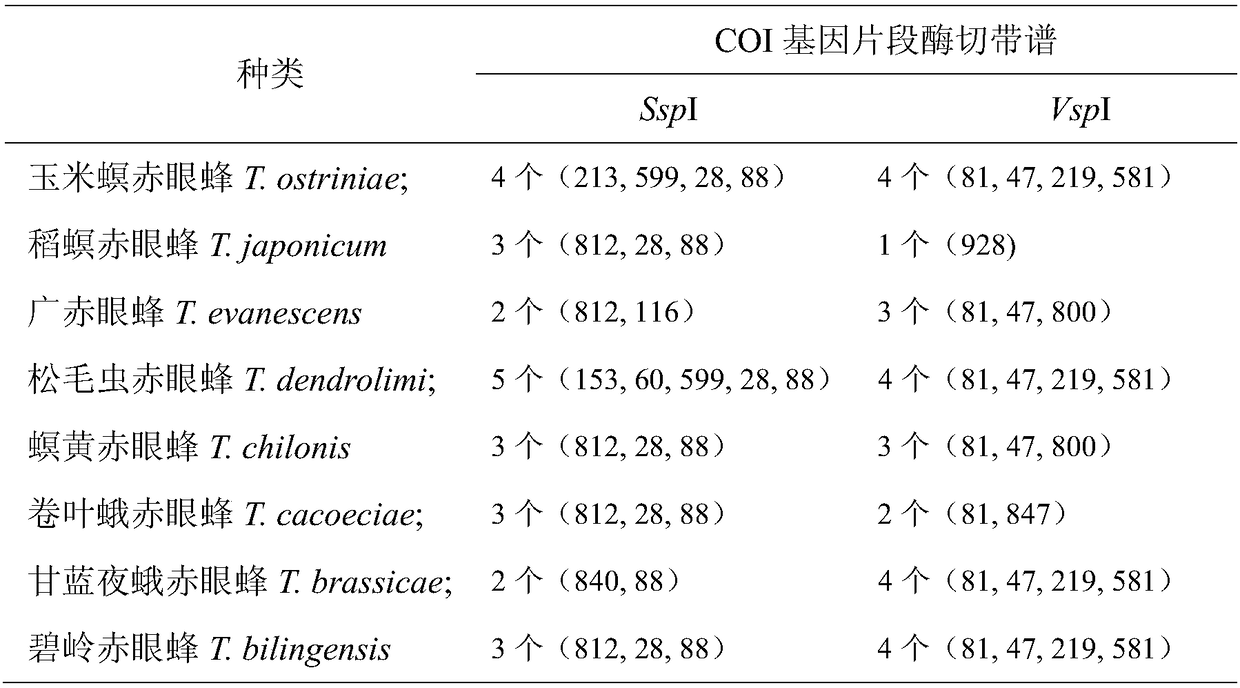
[0032] Example 1 Determination of the SspI standard band spectrum and the VspI enzyme digestion standard band spectrum of 8 species of Trichogramma
[0033] 1 Trichogramma sample
[0034] 8 species of Trichogramma, including Trichogramma brassicae, T.cacoeciae, T.chilonis, T.evanescens, T. japonicum, T. ostriniae, T. bilingensis and T. dendrolimi.
[0035] 2 test hosts
[0036] Rice moth eggs.
[0037] 3 Production of egg cards
[0038] Take fresh rice moth eggs with similar size and shape and good plumpness, use glue to paste single rice moth eggs on coordinate paper to make egg cards (3cm×2cm), equidistant (0.4cm) on one egg card Paste 20 eggs, the degree of paste is easy to peel and not easy to fall off. The prepared egg card is placed in a glass test tube (the specification is 10cm×3cm). At the same time, put a 1cm x 1cm piece of filter paper in the test tube and drip two drops of distilled water to keep it moist, and seal the glass test tube mouth with a absorbent c...
Embodiment 2
[0081] The detection verification of embodiment 2 corn borer Trichogramma
[0082] The host eggs to be tested are the eggs of the Asian corn borer, and the parasitic Trichogramma is T. ostriniae.
[0083] Take the Trichogramma DNA in the eggs of the Asian corn borer to be detected. DNA was extracted using ProMEGA's SVGenomic DNA Purification System Kit. The extraction method is as follows:
[0084] (1) Configure Mix as a buffer for extracting DNA. The buffer system is 275 μl, including Nuclei Lysis Solution (200 μl), 0.5M EDTA (pH8.0) (50 μl), Proteinase K (20 mg / ml) (20 μl), Rnase ASolution (4 mg / ml) (5 μl).
[0085] (2) Dip the mix with a dissecting needle and transfer the host eggs containing unemerged Trichogramma / unparasitized host eggs to a 1.5ml centrifuge tube, add 50μl Mix with a pipette (Gilson, Eppendorf) and use a plastic grinding rod Grind.
[0086](3) After grinding sufficiently, draw 225 μl of Mix to rinse the grinding rod.
[0087] (4) Vortex and mix we...
Embodiment 3
[0099] The detection verification of embodiment 3 rice borer Trichogramma
[0100] The host eggs to be detected are eggs of cotton bollworm, and the parasitic Trichogramma is T. japonicum.
[0101] Other operations are the same as in Example 2.
[0102] The resulting SspI digested products and VspI digested products were detected by agarose gel electrophoresis, and DNAMAN 7.0 software was used to analyze the electrophoresis results. The estimated values of the three bands cut by SspI were 812bp, 28bp, and 88bp, respectively, and the VspI enzyme The estimated value of the excised band is 928bp, and the number and size of the found bands match the standard band of Trichogramma rice borer T. japonicum, confirming that the detection is correct.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com