A novel function of a Bacillus subtilis GGT protein degradation product and antimicrobial peptide identification for the product
A Bacillus subtilis, protein degradation technology, applied in the direction of chemicals for biological control, animal repellents, plant growth regulators, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Purification and mass spectrometry identification of Bacillus subtilis GGT protein.
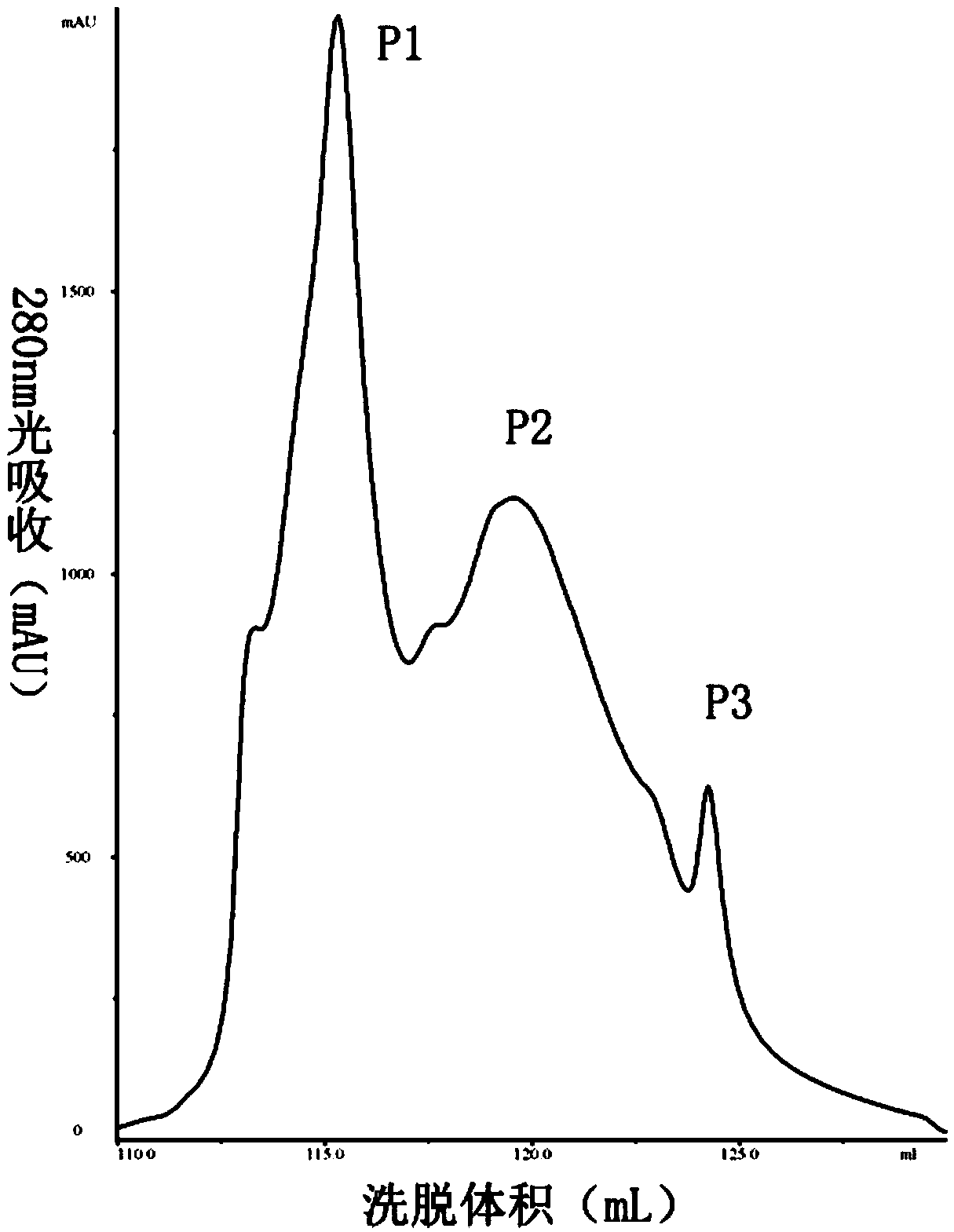
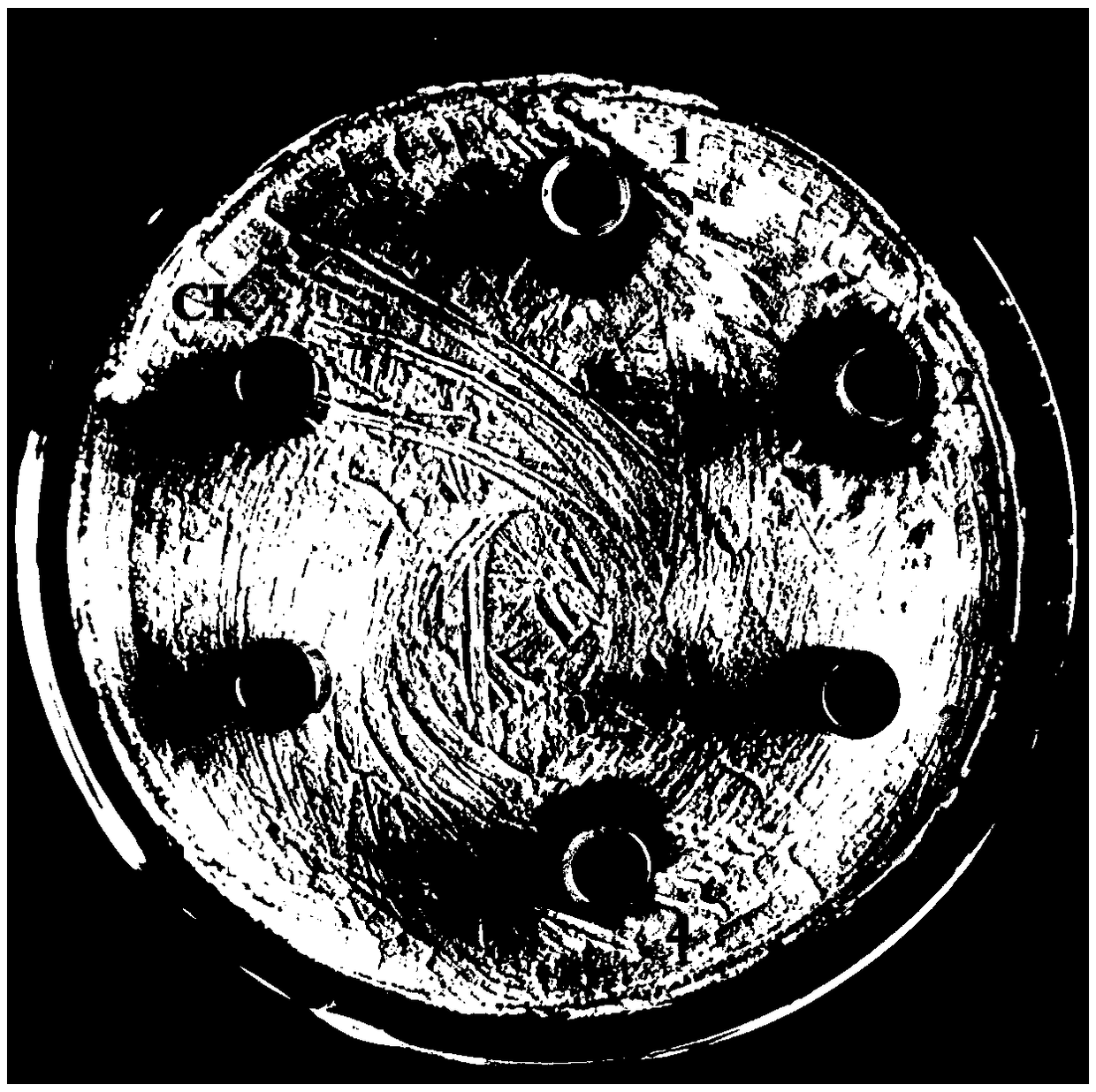
[0035] Inoculate Bacillus subtilis BU412 into YME liquid medium (maltose 10g / L, yeast extract powder 4g / L, glucose 4g / L, pH 7.5) for overnight culture, centrifuge at 12000g to collect the supernatant, and use 80% ammonium sulfate precipitation to obtain For protein components, use 20mM Tris-HCl (pH7.5) buffer to dissolve the precipitate, filter through a 0.22μm filter membrane, and use an AKTA protein purification instrument for anion exchange chromatography. The purification column used is Q hp anion exchange from GE. Column (column volume is 5 mL). Use 20mM Tris-HCl (pH7.5) buffer to load and equilibrate, perform 0-100% linear gradient elution with 0.5M NaCl, 20mM Tris-HCl (pH7.5) buffer, and collect protein according to the absorption peak Components P1-P3 ( figure 1 ), using the Oxford cup method to measure the antibacterial activity of each isolated component to Streptomyces sca...
Embodiment 2
[0038] Prediction of antimicrobial peptides from Bacillus subtilis GGT proteins.
[0039] The amino acid sequence of the GGT protein (Genbank accession number is KIU12867.1) in CAMP R3 Online prediction of the antimicrobial peptide region on the database found that multiple polypeptides of GGT may constitute antimicrobial peptides, combined with the secondary structure and positioning of these polypeptides in the three-dimensional structure of the GGT protein ( Image 6 ), from which 3 polypeptides (Table 1) were selected for online analysis in the APD database. The results showed that all three peptides could form α-helices with hydrophobic surfaces, which may have the activity of antimicrobial peptides. According to the number of amino acids in the three polypeptide chains, they were named LT30, AE26 and IF20, respectively. In addition, the three-dimensional structure of the GGT protein was queried from the PDB database and the three peptide chains were spatially positioned ...
Embodiment 3
[0044] Synthesis and activity detection of predicted antimicrobial peptides.
[0045] Three candidate antibacterial peptides of the GGT protein of Bacillus subtilis were synthesized by solid-phase peptide synthesis technology. The N-terminus of the synthesized peptides was acetylated and the C-terminus was amidated, with a purity of more than 95%. The polypeptide was dissolved in 20 mM Tris-HCl (pH 8.0) buffer at a final concentration of 50 mM. The antibacterial activity of these three polypeptides against Streptomyces scabies was determined by the Oxford cup method. The results showed that polypeptide LT30 had no inhibitory effect on Streptomyces scabica, AE26 and IF20 had inhibitory effect on Streptomyces scabica, and the effect of IF20 was better than that of AE26 ( Figure 7 ).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com