Recombination strain for expressing phospholipase D and application
A recombinant strain and phospholipase technology, applied in the biological field, can solve the problems of gene cloning and expression research, and achieve the effects of good enzyme activity stability, good ability to transfer phosphatidyl groups, and improved enzyme activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] The present invention provides a phospholipase D, comprising 541 amino acids, the amino acid sequence shown in SEQ ID NO.1, the gene encoding the phospholipase D has the nucleotide sequence shown in SEQ ID NO.2, the full length is 1,623 nucleotides.
[0048] The present invention also provides a recombinant plasmid expressing phospholipase D, including the nucleotide sequence shown in SEQ ID NO.2, a signal peptide gene for extracellular expression of phospholipase D, and ribosomes for expressing phospholipase D A binding site, a promoter for expressing phospholipase D, wherein the nucleotide sequence of the signal peptide gene is shown in SEQ ID NO.3, with a full length of 96 nucleotides encoding 32 amino acids; ribosome binding site The nucleotide sequence of the spot (RBS) is shown in SEQ ID NO.4, with a total of 15 nucleotides; the nucleotide sequence of the promoter is shown in SEQ ID NO.5, with a total length of 372 nucleotides.
Embodiment 2
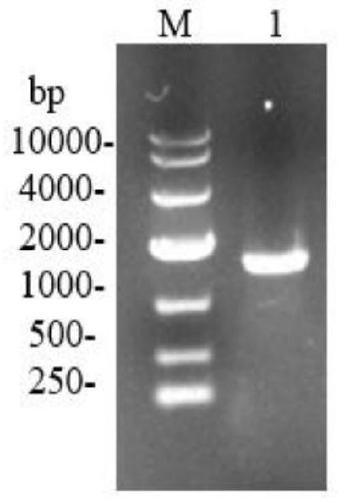
[0050] This embodiment provides the construction of the recombinant plasmid pMA5-pld and its expression method in Bacillus subtilis, the specific steps are as follows:
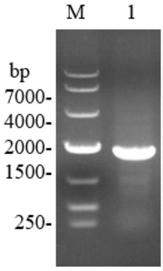
[0051] (1) Amplification of phospholipase D coding sequence
[0052] Using the recombinant plasmid pET-28a(+)-spld containing the phospholipase D coding gene as a template, design primers (P1, P2) to amplify the phospholipase D coding sequence:
[0053] Primer P1: 5'-CG GGATCC ATGGCACGTCATCCGC-3' (BamHI)
[0054] Primer P2: 5'-CG ACGCGT TTAATCCTGACAAATA-3'(MluI)
[0055] The PCR amplification reaction was carried out in a 50 μL system, and 25 μL was added to the reaction system (Premix), 20 μL ddH 2 O, 2 μL template DNA, 1.5 μL each for upstream and downstream primers. The reaction conditions were 30 cycles of denaturation at 94°C for 3 minutes, denaturation at 94°C for 30 s, annealing at 58.4°C for 30 s, extension at 72°C for 1.5 min, and a total of 30 cycles; final extension at 72°C for 10 min. The...
Embodiment 3
[0061] This embodiment provides the construction of the recombinant plasmid pMA5-npld and its expression method in Bacillus subtilis, the specific steps are as follows:
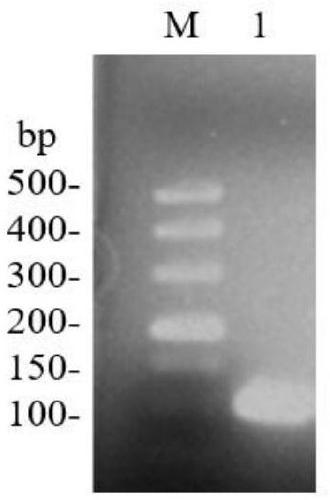
[0062] (1) Amplification of NprB signal peptide sequence
[0063] Using the B. subtilis168 genome as a template, design primers (P3, P4) to amplify the NprB signal peptide sequence:
[0064] Primer P3: 5'-CG GGATCC CGCAACTTGACCAAGAC-3' (BamH I)
[0065] Primer P4: 5'-TTGCGCGGATGACGTGCCATAGCAGCTGAGGCATGTGTTA-3'
[0066] The PCR amplification reaction was carried out in a 50 μL system, and 25 μL was added to the reaction system (Premix), 20 μL ddH 2 O, 2 μL template DNA, 1.5 μL each for upstream and downstream primers. The reaction conditions were 30 cycles of denaturation at 94°C for 3 minutes: denaturation at 94°C for 30 s, annealing at 54.6°C for 30 s, extension at 72°C for 0.5 min, and a total of 30 cycles; final extension at 72°C for 10 min. The PCR products were identified by nucleic acid electrop...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com