Oligo probe and preparation method and application thereof
A probe and reaction technology, applied in the field of Oligo probe and its preparation, can solve the problems of increasing complexity, dependence, and hindering probe preparation, and achieve the effect of low hybridization background, high specificity and high specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0059] HER2, also known as HER2 / neu, c-erbB-2, is a member of the epidermal growth factor receptor (EGFR) family. Studies have shown that HER2 gene amplification exists in more than 30% of human tumors, such as breast cancer, ovarian cancer, Gastric cancer and prostate cancer etc. Breast cancer is a common tumor that threatens women's life and health, and 20%-30% of primary invasive breast cancers have HER2 gene amplification. Fluorescence in situ hybridization (FISH) is a common way to detect HER2 gene amplification.
[0060] (1) Design of the probe
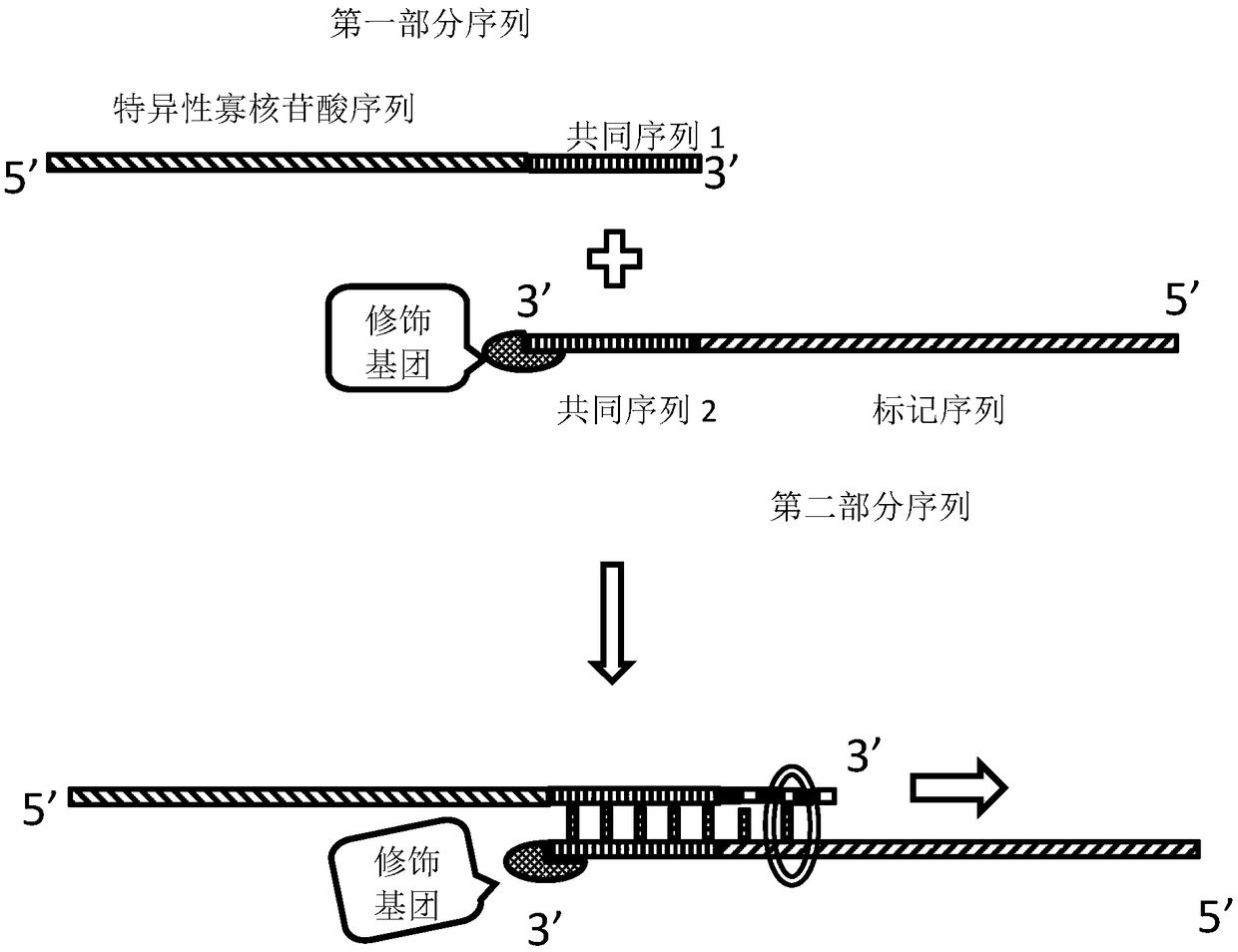
[0061] Find the HER2 gene sequence on chromosome 17 from NCBI, remove the repetitive gene sequence, truncate it into a probe sequence with a length of 50-150bp, and add a common sequence to the 3' end of each sequence (5'—TGGTCGG— 3').
[0062] Probe 1: 5'—ggatccctga tggggagaat gtgaaaattc cagtggccat caaagtgttgTGGTCGG—3';
[0063] Probe 2: 5'—agggaaaaca catcccccaa agccaacaaa gaaatcttag acgtaagcccctccaccctc tcctgctagg TGGTCGG—...
Embodiment 2
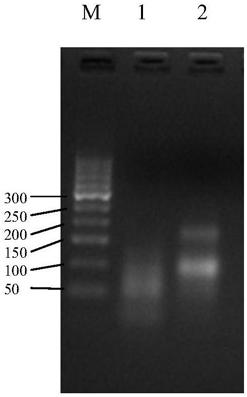
[0095] The above-mentioned labeled HER2 probe (Cy3-labeled HER2 probe) is used for hybridization of cell smears, figure 1 It is a schematic diagram of the labeled probe, which enters the smear cells under the action of the hybridization buffer and specifically binds to the chromosome. The fluorescein combined with the labeled deoxynucleotide emits fluorescence of a specific wavelength under the excitation light. Specific fluorescent signals can be seen after the corresponding grating, such as image 3 , which is the specific bright spot of the excited probe in the cell. This embodiment comprises the steps:
[0096] (1) Preparation of cell smears
[0097] Add colchicine to the culture dish of human embryonic kidney 293T cells cultured in vitro (Shanghai Cell Bank, Chinese Academy of Sciences) to a final concentration of 0.4 μg / mL, and treat at 37°C for 1.5h. Add 25%wt trypsin solution, digest at 37°C for 2min, add 10mL of potassium chloride solution with a concentration of 0...
Embodiment 3
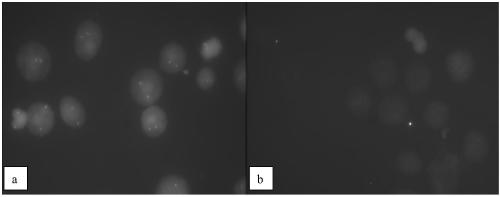
[0107] The 293T cell line in Example 2 was replaced by the HER-2 positive Skbr-3 cell line (Shanghai Cell Bank, Chinese Academy of Sciences), and the cell droplet and hybridization and cleaning methods were the same as in Example 2, and the hybridization results were as follows: Figure 4 , the results showed that the HER2 gene copy number increased significantly in the HER-2 positive Skbr-3 cell line, and the probe prepared by the invention can effectively distinguish the HER-2 positive tumor cell line.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com