Use of tail fiber protein of pseudomonas aeruginosa phage for preparing bacteria detection reagent
A technology of Pseudomonas aeruginosa and tail filament protein, which is applied in the direction of phage, virus/phage, biological testing, etc., can solve the problems of shortening detection time, effort, cross-immunogenicity of antibody batch differences, etc., and achieve rapid detection Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Example 1: Gene recombinant expression of Pseudomonas aeruginosa phage tail filament protein P069
[0041] 1. Materials
[0042] Pseudomonas aeruginosa phage, the strain name is PAP1, and the preservation number is CGMCC No.15569.
[0043] Vector pET-21a (Novagen)
[0044] 2. Method
[0045] 1. Extraction of target gene of Pseudomonas aeruginosa phage tail filament protein P069
[0046] (1) Add 20 μL of 10 μg / μL RNase A and 30 μL of 5 μg / μL DNase I to 950 μL of phage PAP1, and let stand at 37°C for 1 hour;
[0047] (2) Add 1.0mL 40mM EDTA (pH 8.0) to inactivate DNase I;
[0048] (3) Add 5.0 μL of proteinase K and 10 mg of SDS, mix well, and incubate at 56°C for 1 hour to lyse the phage capsid, release DNA, and degrade residual DNase I;
[0049] (4) Add 2.0 mL of balanced phenol (pH 8.0), centrifuge at 5000 g and 4°C for 10 min, and collect the aqueous layer;
[0050] (5) Add 2.0 mL of chloroform, centrifuge at 5000 g and 4°C for 10 min, and collect the aqueous lay...
Embodiment 2
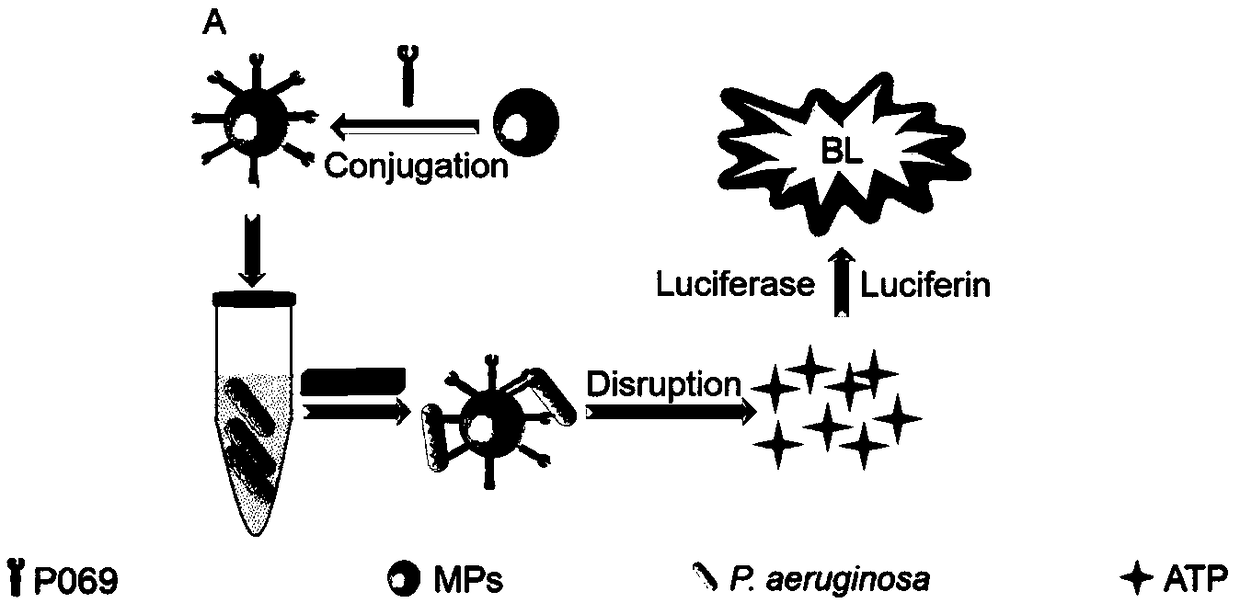
[0073] Example 2. Bioluminescent detection of Pseudomonas aeruginosa based on single site recognition of Pseudomonas aeruginosa phage tail filament protein P069
[0074] 1. Materials
[0075] Pseudomonas aeruginosa phage tail filament protein P069 was prepared according to the method in Example 1.
[0076] Pseudomonas aeruginosa phage, the strain name is PAP1, and the preservation number is CGMCC No.15569.
[0077] Magnetic nanoparticles (Lab on a bead LOABeadsTM AffiAmino, Sweden)
[0078] 2. Method
[0079] 1. Preparation of detection reagents
[0080] (1) Magnetic nanoparticles functionalized with phage tail filament protein P069:
[0081] Take 1.0mL of magnetic particles, wash with 1.0mL of PBST twice; add 1.0mL of PBST and 50μL of activation buffer, react for 15min, then wash with 1.0mL of PBST once, add 1.0mL of 1.0mg / mL React P069 solution at room temperature for 1 hour, then wash twice with PBST, add 80 μL of blocking agent, react at room temperature for 45 minute...
Embodiment 3
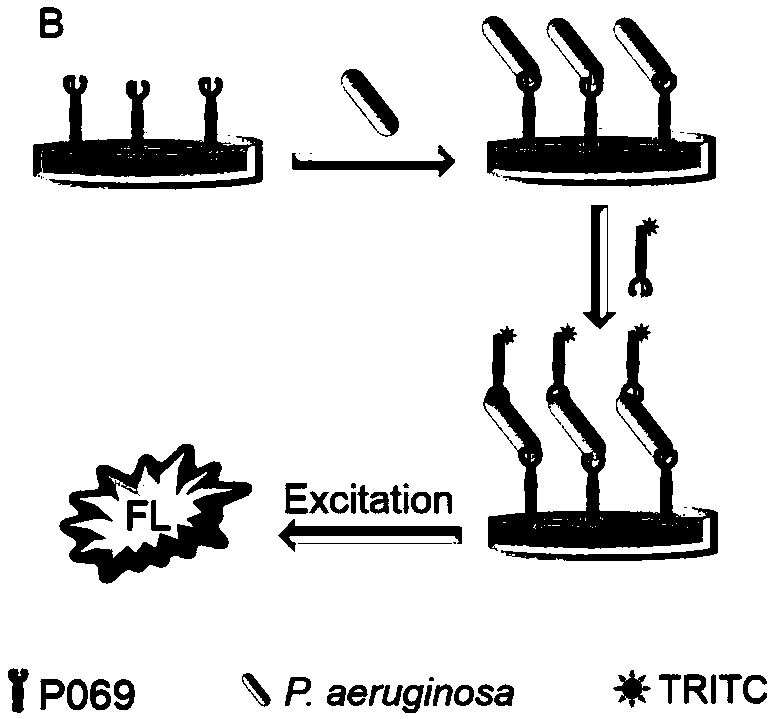
[0098] Example 3. Sandwich fluorescence detection of Pseudomonas aeruginosa based on two-site recognition of Pseudomonas aeruginosa phage tail filament protein P069
[0099] 1. Materials
[0100] Pseudomonas aeruginosa phage tail filament protein P069 was prepared according to the method in Example 1.
[0101] Pseudomonas aeruginosa phage, the strain name is PAP1, and the preservation number is CGMCC No.15569.
[0102] DMSO: purchased from Shanghai Aladdin Biochemical Technology Co., Ltd.
[0103] TRITC: purchased from Shanghai Aladdin Biochemical Technology Co., Ltd.
[0104] 2. Method
[0105] 1. Preparation of detection reagents
[0106] (1) Phage tail filament protein P069: prepared according to the method in Example 1
[0107] (2) Pseudomonas aeruginosa bacterial suspension standard substance:
[0108] Take a single colony of Pseudomonas aeruginosa and inoculate it in LB medium, culture it with shaking at 37°C for 5.5 hours, resuspend the bacteria with Tris buffer (...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com