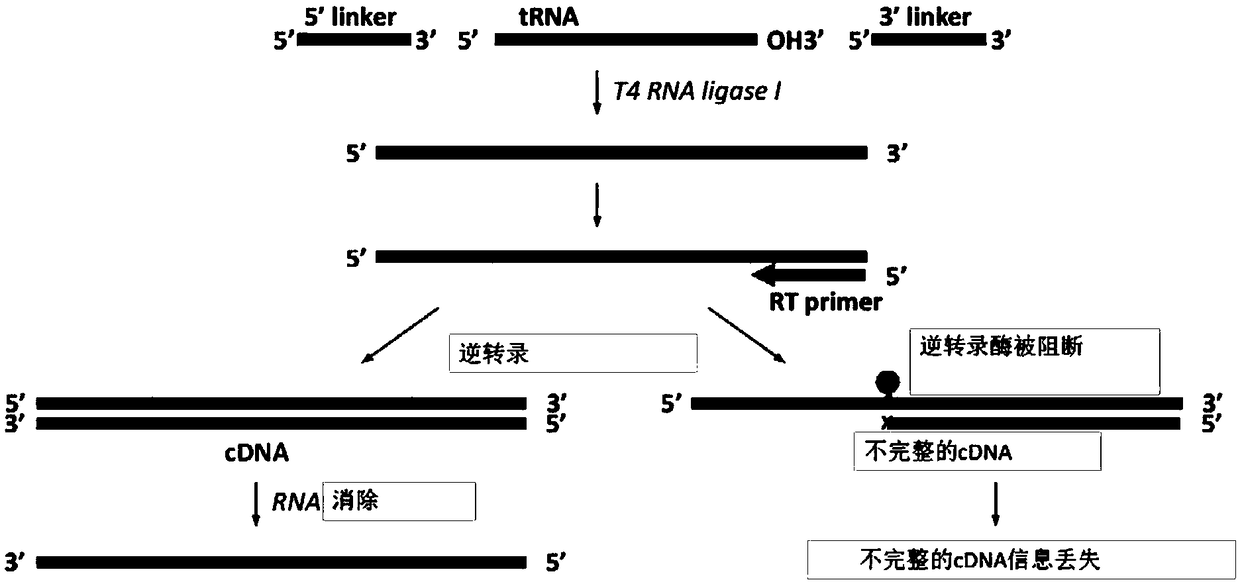
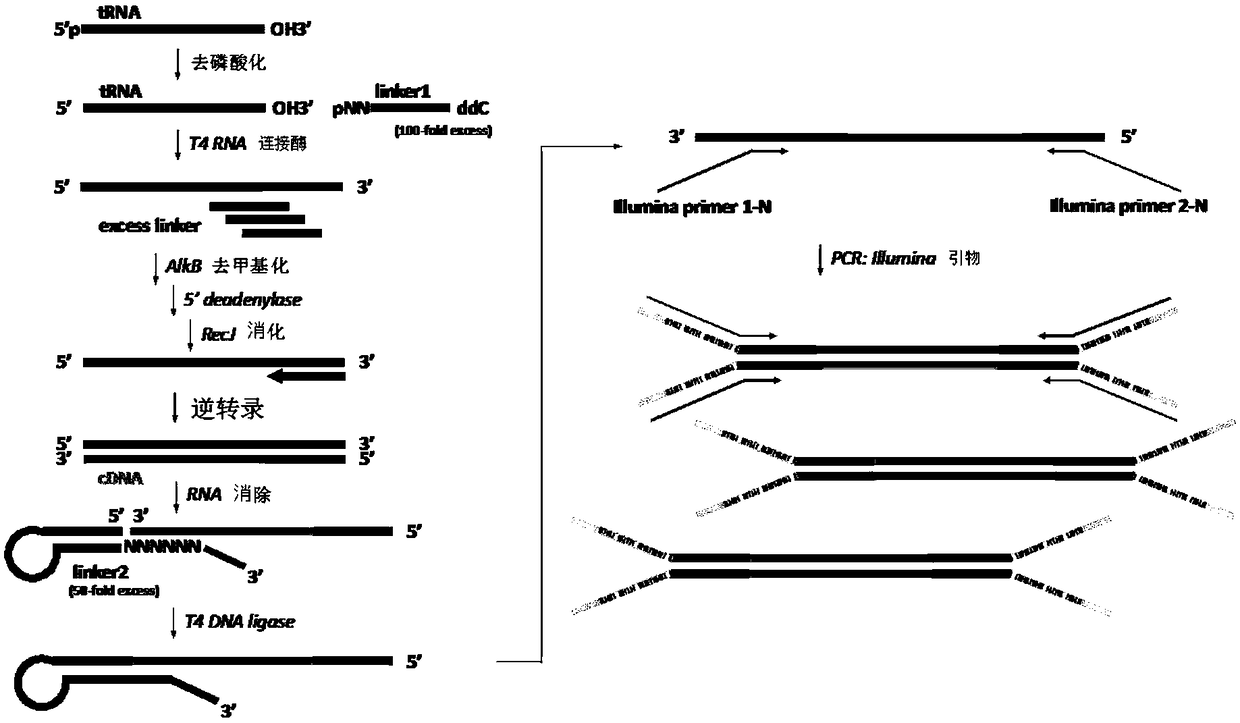
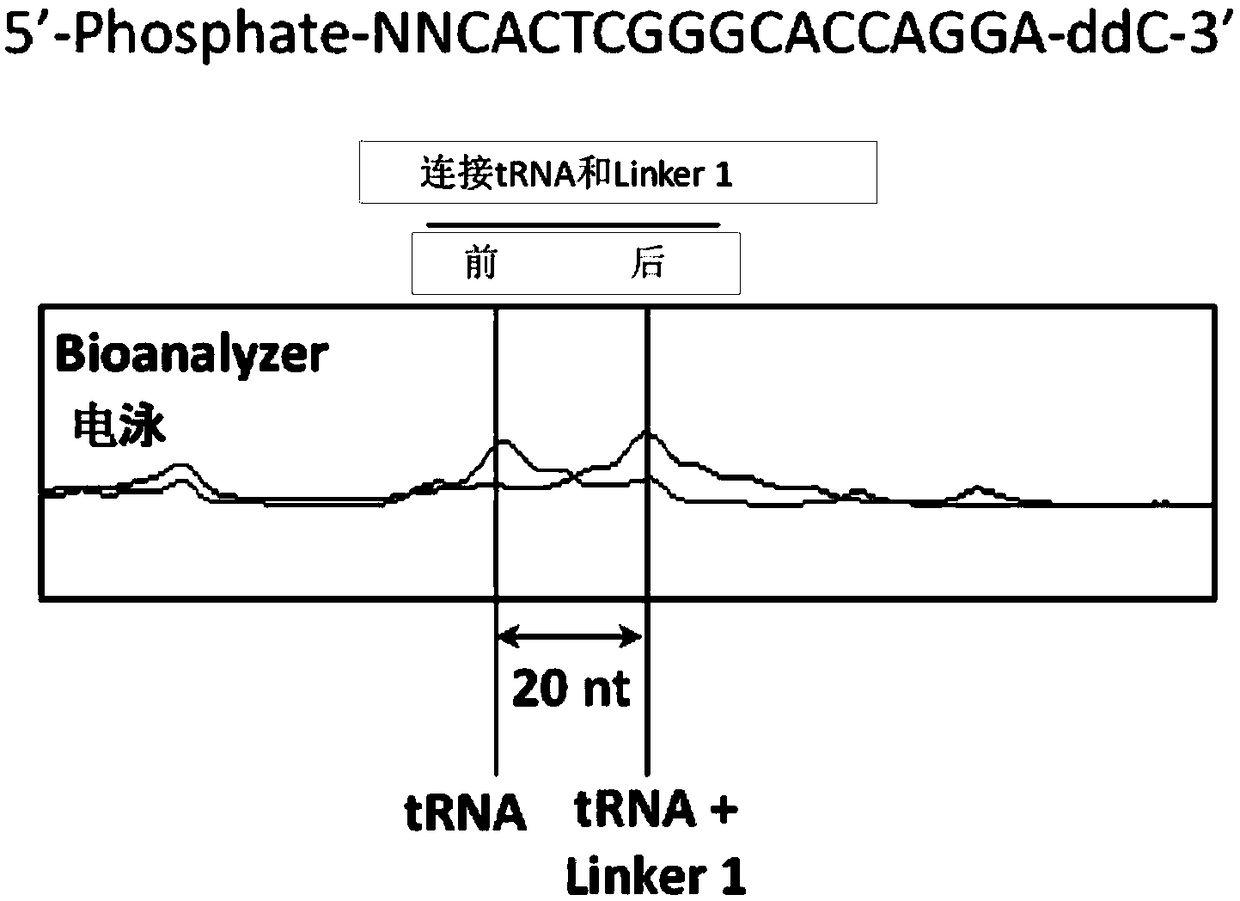
High-throughput sequencing method for absolute quantification of RNA molecules
An absolute quantitative, high-throughput technology, applied in the fields of molecular biology and biomedicine, can solve the problems of ligation efficiency preference that cannot be accurately quantified, RNA sequencing quantification that cannot be modified, etc., and achieve the effect of simple method
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Example 1 Linker1 efficiently connects tRNA samples
[0037] 1. RNA sample preparation
[0038] Monoclonal yeast was inoculated in 5 mL of YPD medium, cultured overnight at 30°C, and 1 mL of bacteria was collected, and RNA samples were extracted with the Purelinkessmall RNA isolation kit (Invitrogen, USA). The operation of the kit was performed according to the instructions.
[0039] 2. Dephosphorylation of RNA samples
[0040] Add 1 μL rSAP (NEB, USA) and 1 μL NEB cutsmart buffer to 50 ng RNA sample, add water to 10 μL, react at 37°C for 15 minutes, and stop the reaction at 65°C for 10 minutes.
[0041] 3. DNA Linker1 connection
[0042]Add 22 μL of the mixed solution (containing 100 pmol Linker1, 10 mmol ATP, 30 units of T4 RNA ligase 1 (NEB, USA), 2 μL buffer, 15 μL PEG8000) to 10 μL dephosphorylated RNA sample. After reacting overnight at 16°C, the reaction solution was purified with the ZymoOligo Clean & Concentrator kit, and the sample was eluted in 19 μL of wa...
Embodiment 2
[0043] Example 2 Linker 2 connects cDNA samples
[0044] 1. DNA sample preparation
[0045] Linker 2 samples were chemically synthesized (IDT, USA), and the sequence is as follows Figure 4 shown. cDNA samples were chemically synthesized DNA samples with a length of 60 nt (IDT, USA), and the sequences were random sequences to simulate cDNA samples from different sources.
[0046] 2. Linker 2 hairpin structure formation
[0047] Dissolve 10 μg Linker 2 in 10 μL water, denature at 95°C for 3 minutes, then cool down to 4°C at a rate of 0.1°C per second to form Figure 4 hairpin structure in .
[0048] 3. Ligation reaction
[0049] The 20 μL reaction system contains: 50 ng cDNA sample, add 1 μg (20 times) or 2.5 μg (50 times) Linker2, 2 μL T4 DNA ligase (NEB, USA), 2 μL buffer, 10 μL PEG8000, 22 ° C reaction After 12 hours, the reaction solution was purified with the ZymoOligo Clean & Concentrator kit, and the sample was eluted in 19 μL of water. bioanalyzer detects connect...
Embodiment 3
[0050] Example 3 Accurate Quantification of tRNA Molecular Changes in the Incubation Period of Mycobacterium tuberculosis
[0051] 1. RNA sample preparation
[0052] Mycobacterium tuberculosis BCG monoclonal cells were inoculated in 100 mL of 7H9 medium (Middlebrook), cultured on a shaker at 37°C for 36 hours, collected and suspended in 100 mL of PBS buffer. Take out 10mL suspension, centrifuge and collect thalli, extract RNA sample, this sample is the sample of incubation period 0th day (S0). The remaining suspension continued to be cultured at 37°C on rollers, and 10 mL of bacteria were taken out on the 4th, 10th, and 20th day of the culture, and the RNA samples were extracted, which were the samples on the 4th (S4), 10th (S10), and 20th (S20) days of the incubation period. Twenty days later, all the thallines were collected, and after 6 days of continuous cultivation with 7H9 medium, the thallines were collected, and the RNA samples were extracted, which was the 6th day (R...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com