Preparation and application of isotope label reagent for analyzing polysaccharides
A technology of isotope labeling and polysaccharides, applied in the field of biomolecular analysis reagents, can solve the problems of lower detection sensitivity and molecular complexity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0174] Embodiment one: QABIT synthesis
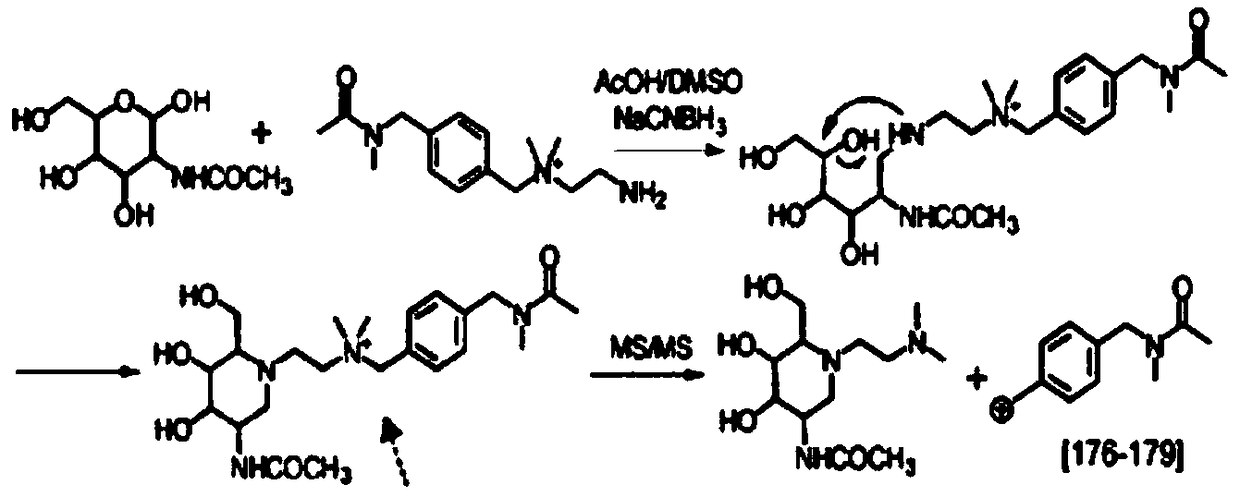
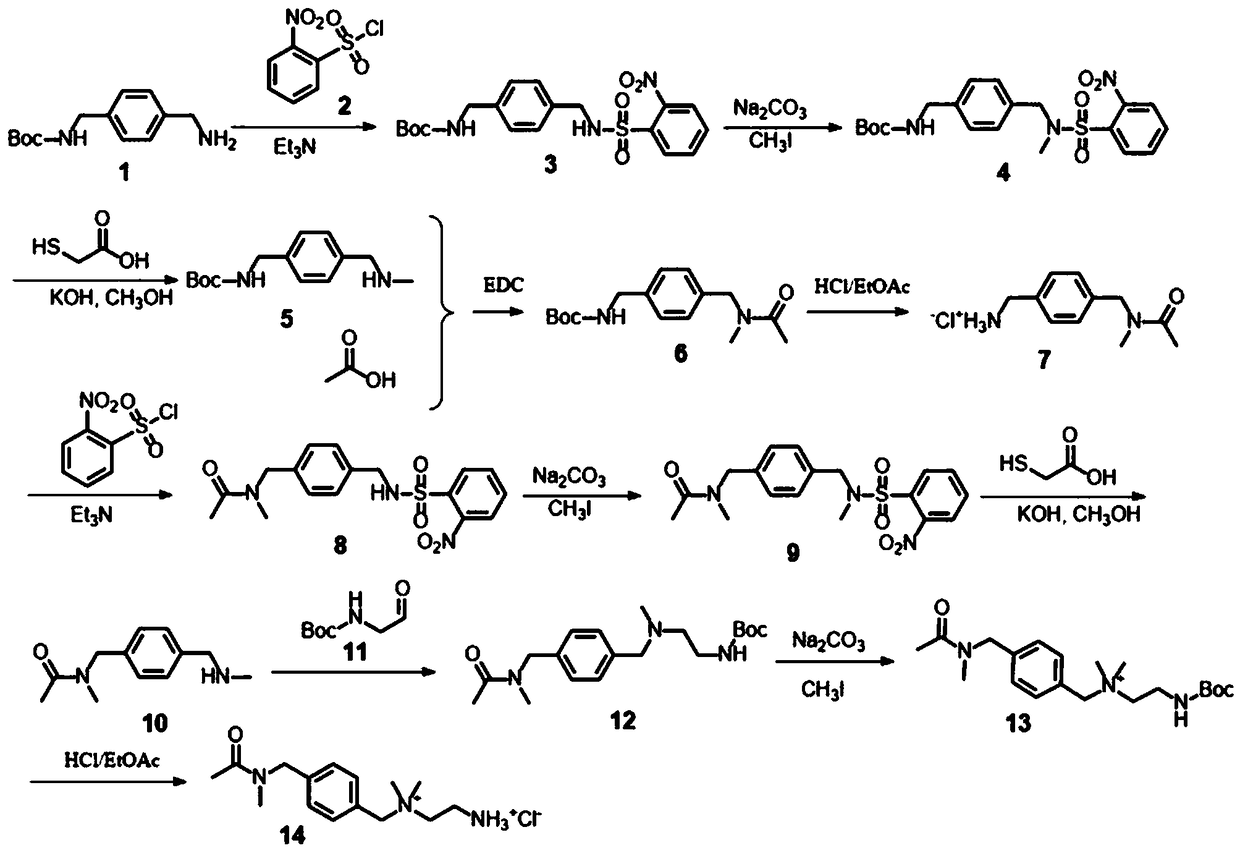
[0175] Such as image 3 The synthetic route of described QABIT; Adopt following concrete synthetic route:
[0176] Compound 1 (1.12 g, 4.75 mmol) and 1.5 mL of triethylamine (10.8 mmol) were mixed in 30 mL of dichloromethane (DCM), then compound 2 (1 g, 4.52 mmol) was added. The reaction was stirred overnight under an atmosphere of argon. After the reaction was complete, 30 mL of DCM was added. The mixture was washed twice with 50 mL HCl (50 mmol / L) solution, twice with 50 mL saturated NaHCO3 solution, and once with 50 mL brine solution. The organic layer was dried over anhydrous Na2SO4 and removed by Rotavap to give 1.7 g of white solid compound 3 (4.03 mmol, 89% yield). 1H-NMR(CDCl3,400MHz)δ7.60-8.00(m,4H), 7.10-7.20(m,4H),4.28(s,2H),4.22(d,J=6.0Hz,2H),1.45(s ,9H).ESI-MS (M+H)+=422.14, Cal. (M+H)+=422.14.
[0177] Compound 3 (1.7 g, 4.40 mmol) was dissolved in 10 mL of DMF, and 2.8 g of Na2CO3 (26.4 mmol) was added as a solid. ...
Embodiment 2
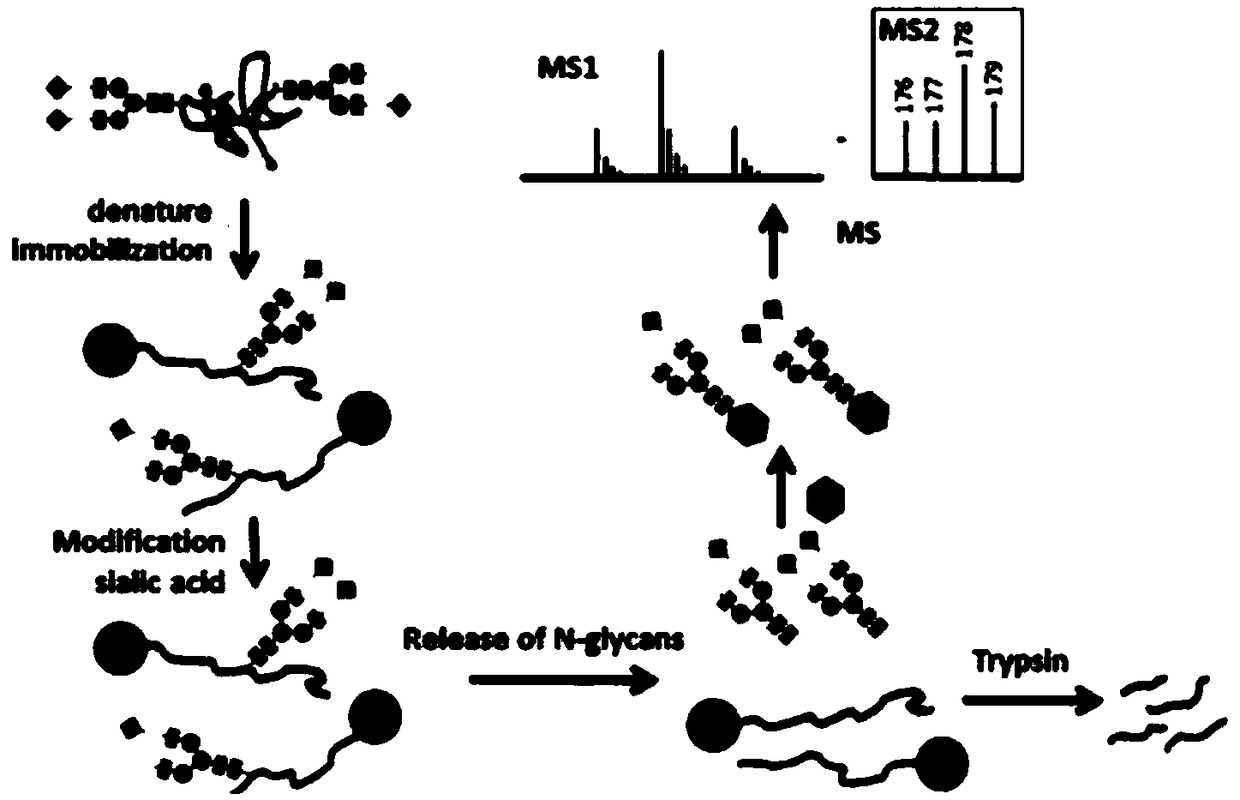
[0188] Example 2: Serum polysaccharide labeling method
[0189] In the first step, N-glycan enrichment.
[0190] First denature 20 μL of serum proteins in 200 μL of a solution consisting of 20 μL of denaturing buffer (10x) and 160 μL of buffer (pH 10.0; 40 mmol / L sodium citrate and 20 mmol / L sodium carbonate) at 100 °C for 10 min. After pre-treating Aminolink™ resin (200 μL) with pH 10 buffer, denatured protein was added to Aminolink™ resin in 300 μL buffer (pH 10.0) and incubated with mixing for 4 hours at room temperature. Add 50 μL of 500 mmol / L sodium cyanoborohydride (1×PBS) and incubate for an additional 4 hours. After rinsing the resin twice with 500 µL of 1x PBS (pH 7.4), the samples were reduced with 50 mmol / L sodium cyanoborohydride (NaCNBH3) in 1x PBS for 4 hours.
[0191] The protein-conjugated beads were washed twice with 1 mol / L Tris-HCl (500 μL, pH 7.6), and the remaining aldehyde sites were washed with 500 μL 1 mol / L Tris-HCl in 50 mmol / L NaCNBH3 (0.5 hr). T...
Embodiment 3
[0197] Example 3: Data analysis of QABIT tags.
[0198]Like other isobaric tags for peptides and small molecules, the 4-fold QABIT reagent tested is a set of four molecules with the same chemical structure and molecular weight, but they contain different stable isotopic nuclei, such as 13C and 2H. Their structure consists of a series of reporter molecules with molecular weights ranging from 176 to 179 Daltons, a balance compensating for differences in reporter molecular masses, and reactive primary amines coupled to polysaccharides by reductive amination. The labeling chemistry is similar to that used by 2-AA / 2-AB (2-aminobenzoic acid (2-AA) or 2-aminobenzamide (2-AB)), Bigge, JC et al. "Nonselective and efficient fluorescent labeling 2 -aminobenzamide and anthranilic acid, Anal.Biochem.230,229-238 (1995)), so the well-known scheme for 2-AA / 2-AB labeling can be modified without many changes in this in the field.
[0199] However, a significant difference between the tags of ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com