Cat serum amyloid protein A and preparation method and application of monoclonal antibody of cat serum amyloid protein A
A serum starch and protein technology, applied in the field of bioengineering, can solve problems such as being unsuitable for rapid diagnosis, easy to delay treatment, cumbersome operation, etc., and achieve the effects of improving detection sensitivity, eliminating interference, and improving expression level.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
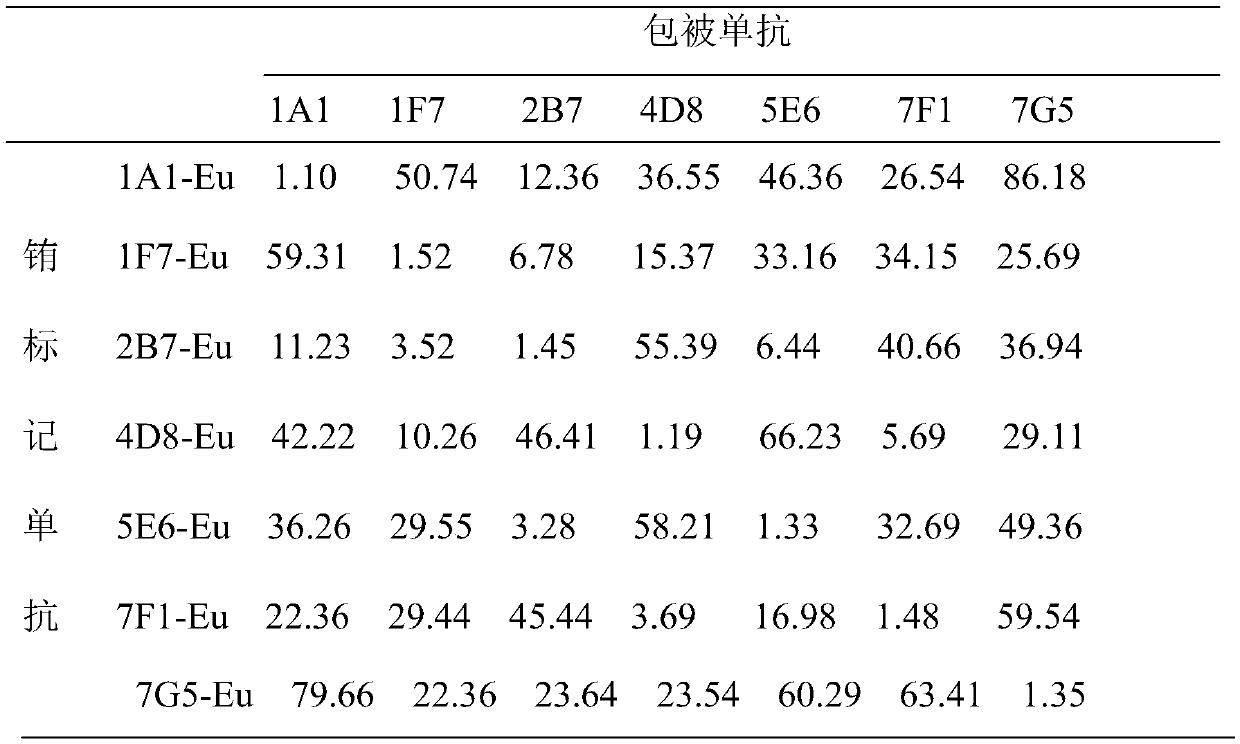
Image
Examples
Embodiment 1
[0009] Example 1: Selection of Predominant Antigen Epitope of Feline Serum Amyloid A
[0010] Taking feline serum amyloid A as the target antigen, using the biological software DNAssist2.0 to analyze the hydrophilicity and antigenicity of the epitope sequence, and selecting the A dominant epitope (SEQ ID No: 2) and the B dominant antigenic epitope (SEQ ID No: 3). At the same time, the sequence comparison results showed that the selected two dominant antigenic epitopes A and B have high sequence specificity and no obvious homology with other protein sequences.
Embodiment 2
[0011] Example 2: Cat Serum Amyloid A Predominant Antigen Epitope Concatenation
[0012] In order to enhance the stimulation of the selected antigenic epitope on the immune system of mice and facilitate subsequent experiments, the two dominant antigenic epitope sequences of cat serum amyloid A, A and B, were connected by flexible fragments (four consecutive glycines). Repeat four more times to obtain the amino acid sequence of the recombinant protein, the specific sequence of which is shown in SEQ ID No: 1 in the sequence table.
Embodiment 3
[0013] Embodiment 3: optimize the nucleotide sequence of coding recombinant protein
[0014] In order to improve the expression of recombinant protein in Escherichia coli, under the premise that the amino acid sequence of the recombinant protein remains unchanged, the amino acid sequence encoding the recombinant protein is converted into the corresponding nucleotide sequence according to the preferred codons of Escherichia coli. The specific sequence is shown in the sequence table Shown in SEQ ID No: 4, and the nucleotide sequences corresponding to the restriction sites BamHI and EcoRI were added in the upstream and downstream, respectively, and synthesized by Hangzhou Xianzhi Biotechnology Co., Ltd. The synthesized target gene was cloned into the pMD19-T vector (Bao Bioengineering Dalian Co., Ltd.).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com