Cell one-step real-time quantitative PCR method, related reagent kit and applications
A real-time quantitative and kit technology, applied in the field of PCR detection, can solve the problems of small safety hazards and low operation difficulty, and achieve the effect of improving efficiency and strong consistency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Embodiment 1 The direct cell lysing method of the present invention and the comparison of traditional RNA extraction method
[0042] 1.1 Cell direct lysis qPCR detection
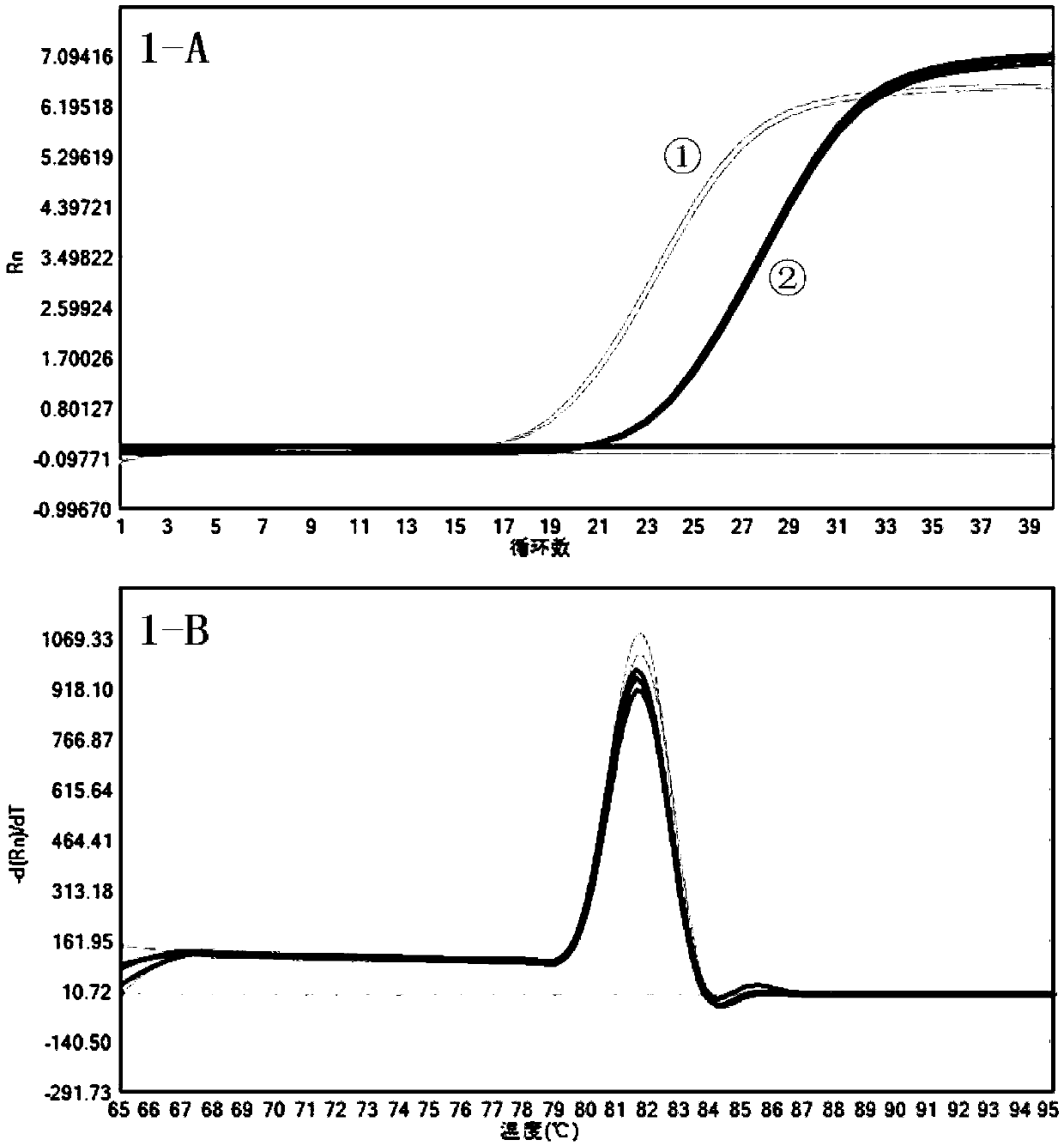
[0043]Prepare 1× lysis buffer with DEPC water: 5mM Tris-Hcl (pH 8.0) + 1-20mM EDTA + 0.1%-2% Triton X-100 + 0.2-2U RNase Inhibitor (available from Wujiang Jinan Protein Technology Co., Ltd.) +0.1M DTT. Resuscitate CHO-DG44 cells and subculture for 3 times. After cell counting, the cells were transferred to EP tubes, and the supernatant was discarded after centrifugation. Add 1× Lysis Buffer (the final cell density is 300 cells / ul); after mixing, place at room temperature for 5-10 minutes, and centrifuge to remove cell debris. The Primer 5.0 software was used to design the intron-spanning primers for the reference gene gapdh in CHO-DG44 cells, and the specificity was detected by Blast. Take 1ul supernatant to 8 tubes, add 2ul 1st Strand cDNASynthesis SuperMix (available from Wujiang Nearshore Pro...
Embodiment 2
[0052] Embodiment 2 direct lysing method cell number detection range and the establishment of standard curve
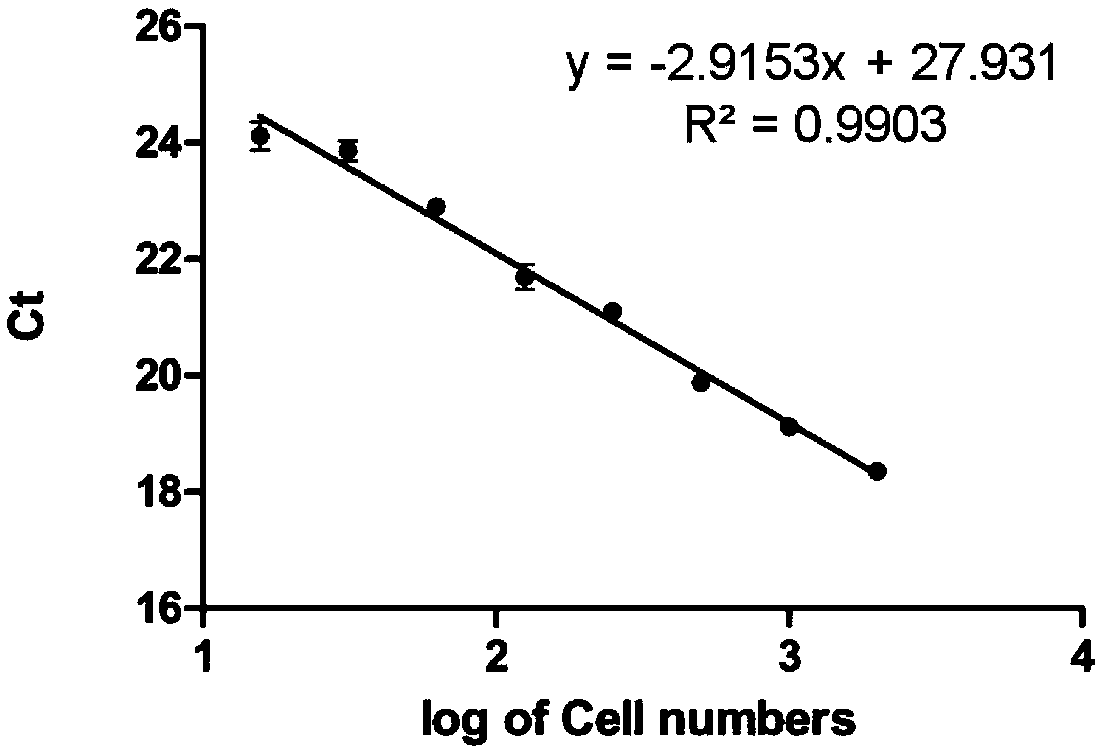
[0053] Cells with a density of 2000 cells / ul were taken and diluted logarithmically with 1×PBS, and the dilution densities were as follows: 1000, 500, 250, 125, 62.5, 31.25, 15.625 cells / ul. Add 10× lysis buffer to lyse at room temperature for 5 minutes, and the supernatant after high-speed centrifugation is used for one-step RT-qCPR detection. The RT-qPCR program is detailed in Table 2. See Table 1 for primer sequences.
[0054] Such as figure 2 The standard curve of the direct cell lysis method shown, where the abscissa is log 10 (number of cells), the ordinate is the Ct value.
[0055] The results of this experiment show that 15-2000 cell lysates can be used for RT-qPCR detection and log 10 (Number of cells) has a linear relationship with the Ct value. It proves that the direct lysis method can detect the expression level of the target gene in a wide range of...
Embodiment 3
[0056] Example 3 Detection of the activating effect of blinatumomab on Jurkat cells by direct lysis method
[0057] 3.1 Cell plating and antibody activation
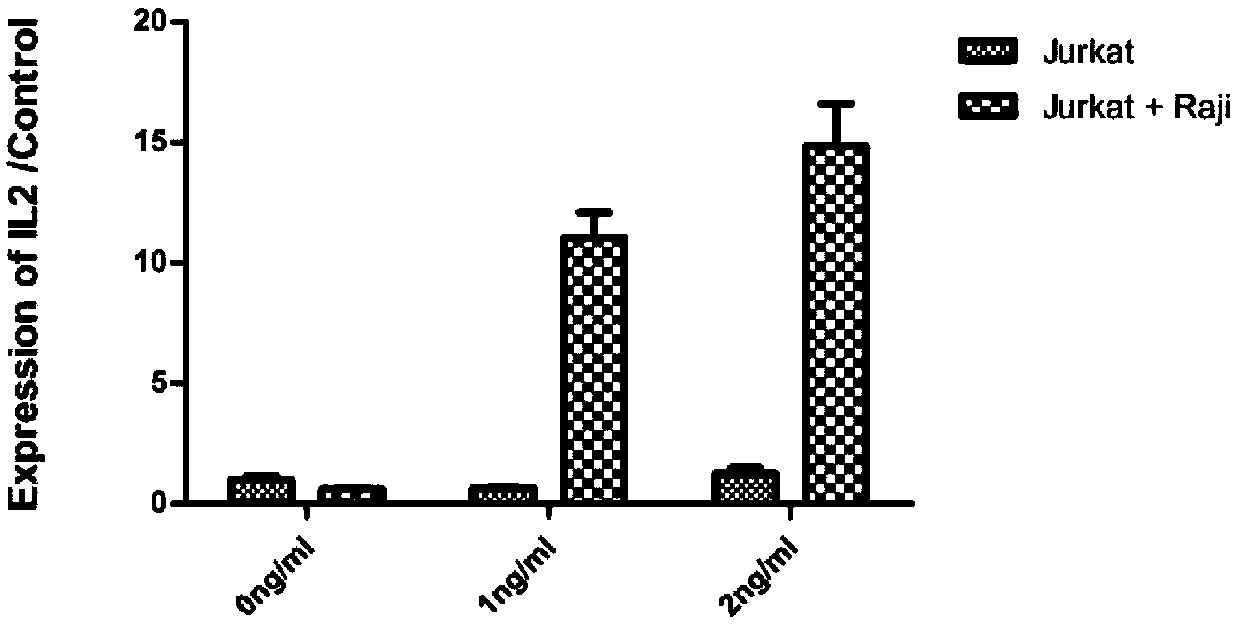
[0058] Jurkat cells were counted, and the cells were centrifuged and resuspended at 0.3×10 6 96-well plate at density per ml; Raji cells were evenly pipetted and counted at 0.3×10 5 cells / ml density into wells pre-coated with Jurkat. Plate overnight, add different concentrations of blinatumomab (purchased from Amgen) antibodies, and incubate overnight.
[0059] 3.2 Detection of activation of blinatumomab antibody by direct lysis method
[0060] After the cells were evenly pipetted, the suspension was pipetted into an EP tube, and 1 / 10 volume of 10× lysis buffer was added. Lyse at room temperature for 5 minutes, and take the supernatant after high-speed centrifugation for one-step RT-qPCR detection. Primer 5.0 software was used to design il-2 and the reference gene gapdh, and Blast was used for specific detection. T...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap