Separated recombinant oncolytic adenovirus, drug composition and application of separated recombinant oncolytic adenovirus to drug for treating tumor and/or cancer
An oncolytic adenovirus and composition technology, applied in the biological field, can solve the problems of T cell anergy or specific immune tolerance and apoptosis, and achieve enhanced anti-tumor immune stimulation, improved safety, and low toxicity Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example 1
[0110] Preparation Example 1: Construction of E1A Gene Expression Vector
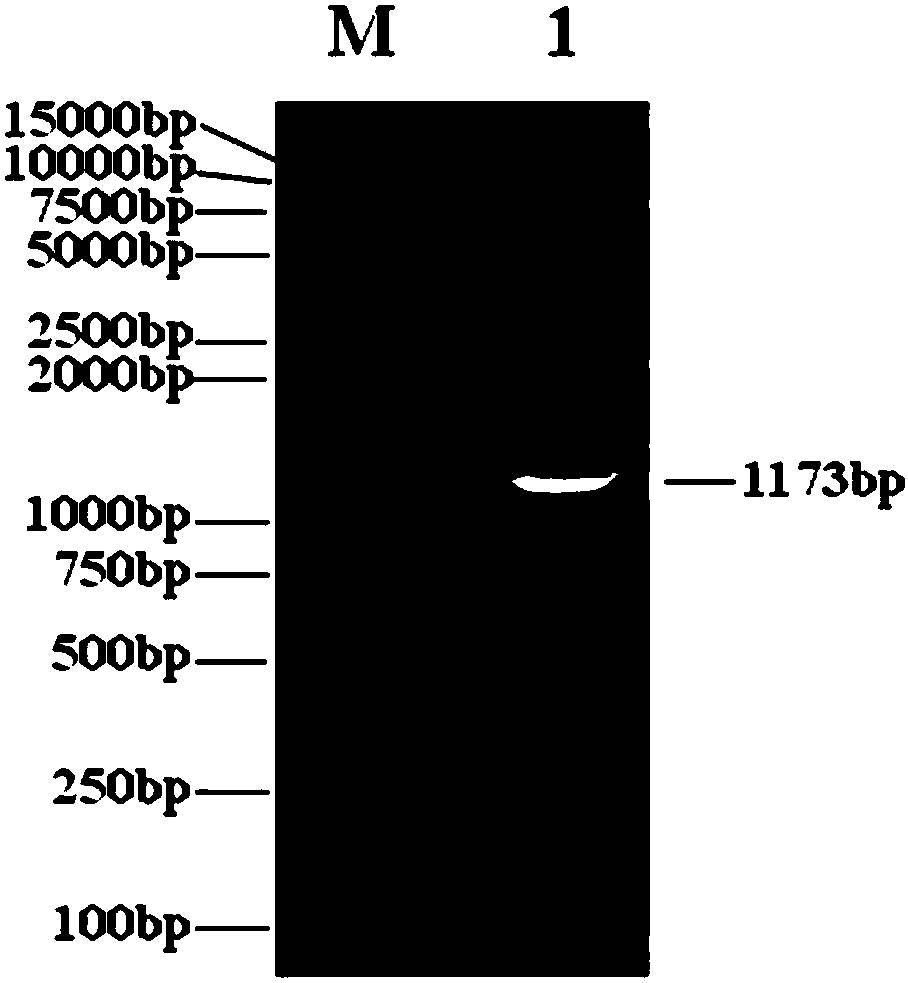
[0111] According to the human adenovirus type 5 (AD5) genome DNA sequence (ACCESSION: AC_000008) in the Genbank of NCBI (that is, the National Center for Biotechnology Information, website: https: / / www.ncbi.nlm.nih.gov) design two PCR primers (P1: GGA AGATCTGGACTGAAAATGAG (SEQ ID No.1), P2: TGAGGTCAGATGTAACCAAGATTA (SEQ ID No.2); note: the 5' end of the primer P1 has added a BglII restriction site, underlined); extracted from Shanghai Sanwei Biotechnology Co., Ltd. Oncolytic virus (H101) genomic DNA was used as a template for high-fidelity PCR amplification of the 1164bp sequence between 551-1714 on the AD5 genomic DNA, and the actual size was 1173bp (see figure 1 ), this sequence includes the coding region of the E1A gene (excluding the E1A promoter sequence) and part of the 3'UTR region. Utilize the PCR product that BglII digestion obtains, and it is cloned between the BglII and the EcoRV site in t...
preparation example 2
[0114] Preparation example 2: construction of shRNA expression vector
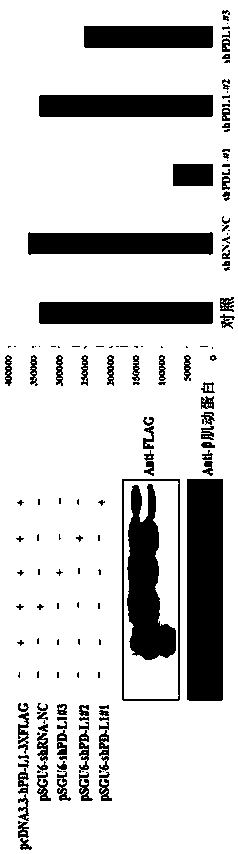
[0115] According to the human PDL1variant1 sequence (ACCESSION: NM_014143) in Genbank on the NCBI website, three shRNA sequences (shPDL1-1 (or shPDL1-#1), shPDL1-2 (or shPDL1-#2) and shPDL1-3 (or shPDL1-#3), respectively SEQ ID NO.16, SEQ ID NO.19, SEQ ID NO.22) In addition, a negative control sequence shPDL1-NC that has nothing to do with human PDL1 mRNA was also designed. The sequence is as follows:
[0116] (1) shPDL1-1
[0117] Synthetic sense sequence (SEQ ID No.14):
[0118] 5'-CACC GGGAAATGGAGGATAAGAACA TGTTTCTTATCCTCCATTTCCC -3'
[0119] Synthetic antisense sequence (SEQ ID No.15):
[0120] 5'-AGCT GGGAAATGGAGGATAAGAACA TGTTCTTATCCTCCATTTCC C- 3'
[0121] shRNA DNA (SEQ ID No.16):
[0122] GGGAAATGGAGGATAAGAACA TGTTTCTTATCCTCCATTTCCC
[0123] (2) shPDL1-2
[0124] Synthetic sense sequence (SEQ ID No.17):
[0125] 5'-CACC GGATCCAGTCACCTCTGAACA TGTTCAGAGGTGAC...
preparation example 3
[0147] Preparation Example 3: Preparation of Genomic DNA of Oncolytic Adenovirus (OAd-shPDL1)
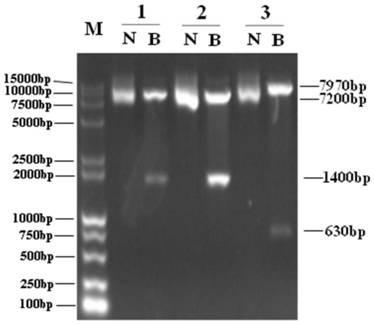
[0148] 1. After confirming the inhibitory effect of shPDL1, clone the coding frame including the U6 promoter and the entire sequence of shPDL1 into pShuttle-MCS-E1A. First, use SacI to digest the pSGU6 / GFP / Neo-shPDL1 vector, then use T4 DNA polymerase to cut the sticky ends formed after SacI digestion, and then carry out ethanol / ammonium acetate precipitation and recovery, and finally digest with KpnI and recover including Coding frame sequence including the U6 promoter and shPDL1 sequence; at the same time, the pShuttle-MCS-E1A vector was digested with KpnI and EcoRV and recovered, and finally the three shPDL1 coding frames were respectively connected to the pShuttle-MCS-E1A vector to obtain the final Vector pShuttle-U6-shPDL1-CMV-E1A, see the procedure Figure 10 . Select a few colonies to extract the plasmid and carry out KpnI / HindIII enzyme digestion identification, and the co...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com