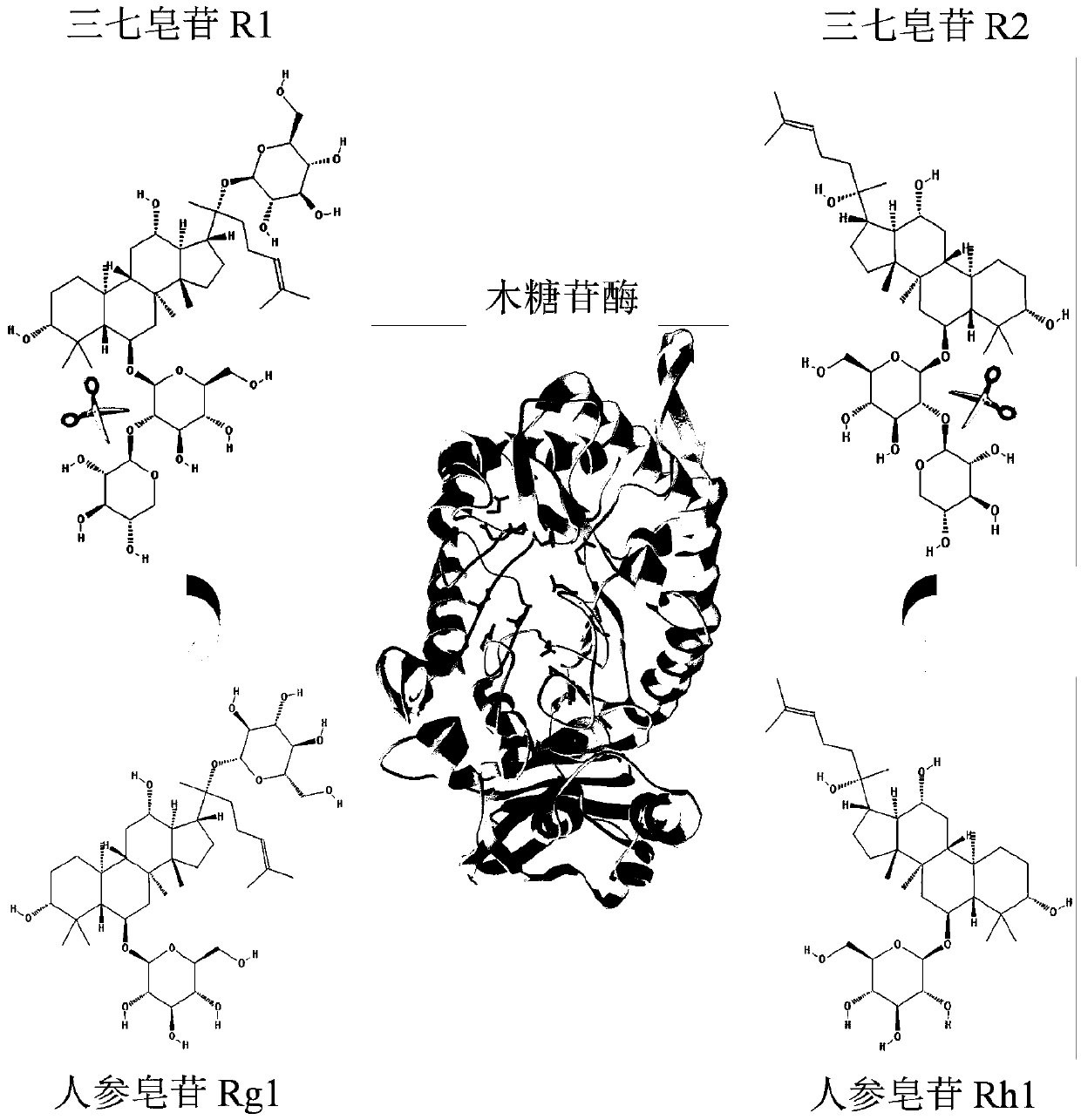
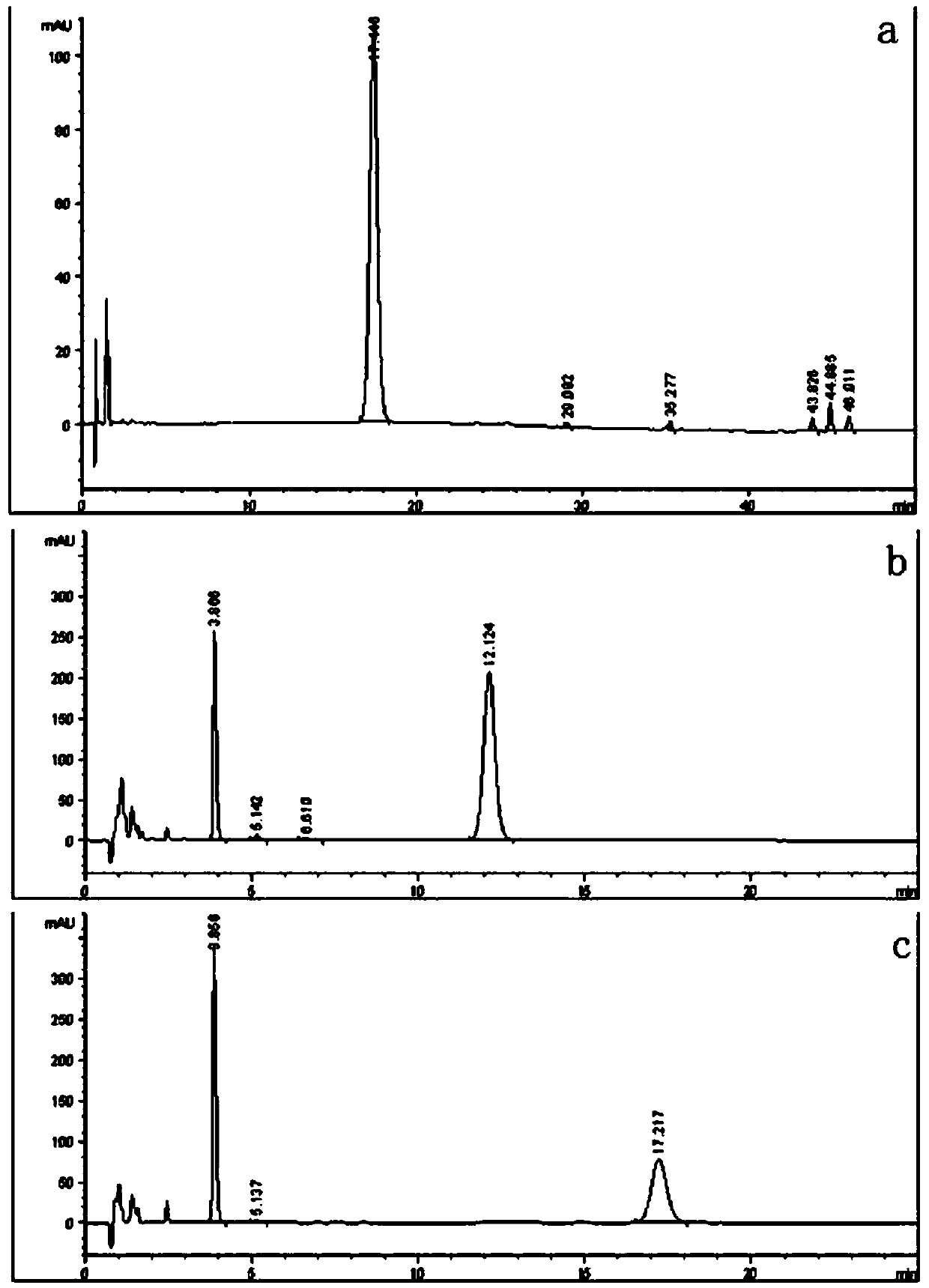
Xylosidase mutant capable of transforming notoginsenoside R1 into ginsenoside Rg1
A technology of xylosidase and ginsenoside, which is applied in the field of genetic engineering and can solve problems that are not conducive to the purification of ginsenoside Rg1
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Construction and transformation of embodiment 1 wild enzyme JB13GH39 expression vector
[0034] 1) Extraction of Sphingomonas Genomic DNA
[0035] Centrifuge the liquid bacterial solution cultured for 2 days to get the bacterial cells, add 1mL lysozyme, treat at 37°C for 60min, then add the lysate, the composition of the lyse is: 50mM Tris, 20mM EDTA, NaCl 500mM, 2% SDS (w / v), pH8.0, lysed in 70°C water bath for 60min, mixed every 10min, centrifuged at 10000rpm at 4°C for 5min. The supernatant was extracted in phenol / chloroform to remove impurity proteins, and then an equal volume of isopropanol was added to the supernatant. After standing at room temperature for 5 minutes, centrifuge at 10,000 rpm for 10 minutes at 4°C. The supernatant was discarded, the precipitate was washed twice with 70% ethanol, dried in vacuum, dissolved by adding an appropriate amount of TE, and stored at -20°C for later use.
[0036] 2) Amplify or gene synthesize the coding sequence of wild e...
Embodiment 2
[0039] Construction and transformation of embodiment 2 mutant enzyme MutY257T expression vector
[0040] Using the recombinant plasmid pEasy-E2-jB13GH39 as a template, according to the mutation kit Mut II Fast Mutagenesis Kit V2 method for mutation and recombination, the mutation primers are 5'ACAGCaccGGCGTCGATGGCGGCTTTCTCGAC 3' and 5'ATCGACGCCggtGCTGTGCGTGGTGACGAAAT 3', the PCR reaction parameters are: denaturation at 94°C for 5min; then denaturation at 94°C for 30sec, annealing at 60°C for 30sec, and extension at 72°C 3min, after 30 cycles, keep warm at 72°C for 10min.
[0041] As a result, the recombinant plasmid pEasy-E2-MutY257T containing the MutY257T coding gene was obtained, and the plasmid pEasy-E2-MutY257T was used to transform Escherichia coli BL21(DE3) to obtain the recombinant strain BL21(DE3) / MutY257T containing the MutY257T coding gene.
Embodiment 3
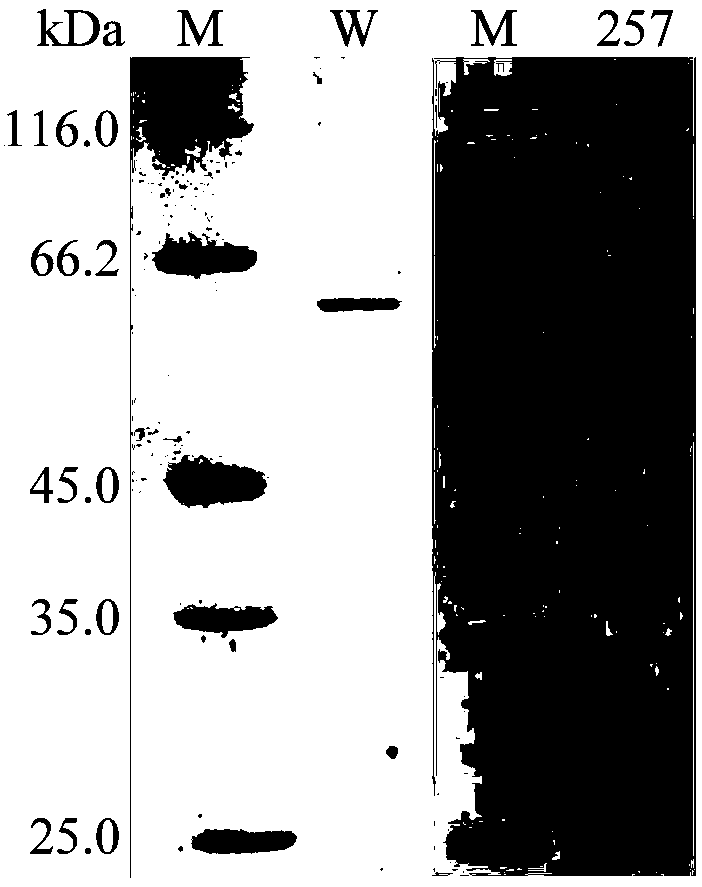
[0042] Embodiment 3 Preparation of wild enzyme JB13GH39 and mutant enzyme MutY257T
[0043] The recombinant strains BL21(DE3) / jB13GH39 and BL21(DE3) / MutY257T were inoculated in LB (containing 100 μg mL -1 Amp) medium, shake rapidly at 37°C for 16h.
[0044] Then the activated bacterial solution was inoculated into fresh LB (containing 100 μg mL -1 Amp) culture medium, rapid shaking culture for about 2–3h (OD 600 After reaching 0.6–1.0), add IPTG with a final concentration of 0.7mM for induction, and continue shaking culture at 20°C for about 20h or 26°C for about 8h. Centrifuge at 12000rpm for 5min to collect the bacteria. After suspending the cells with an appropriate amount of McIlvaine buffer solution with pH 7.0, the cells were ultrasonically disrupted in a low-temperature water bath. After the crude enzyme solution concentrated in the cells was centrifuged at 12,000rpm for 10min, the supernatant was aspirated and the target protein was affinity and eluted with Nicke...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com