Illumina sequencing technology based kit and method for detecting hepatitis B typing and multi-drug resistant sites
A technology for hepatitis B and technical detection, applied in the biological field, can solve problems such as the difficulty in detecting low-titer HBV drug-resistant mutations, and achieve stable amplification effects, strong pertinence, and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
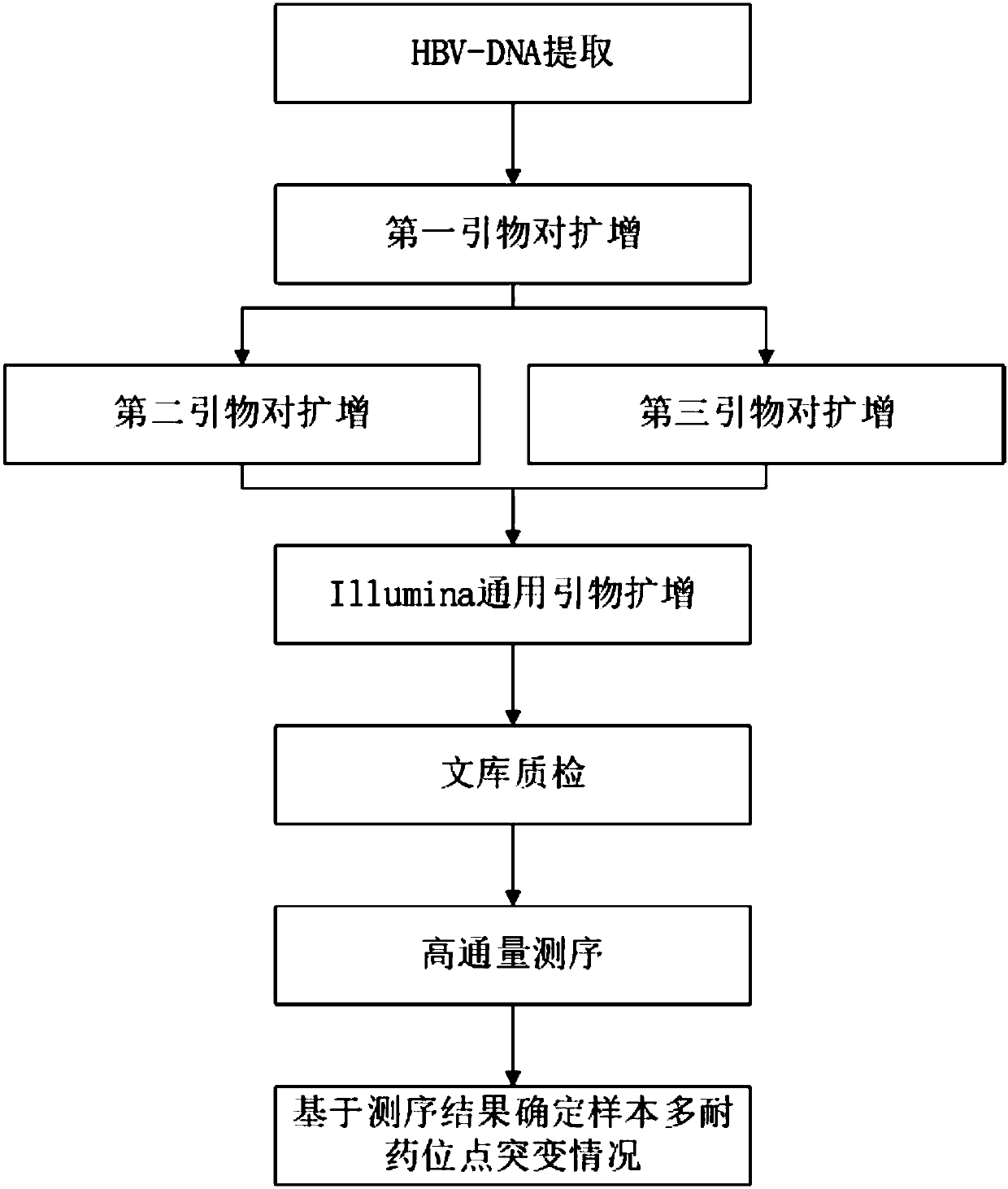
[0079] In this embodiment, Illumina sequencing technology is used to detect HBV samples in patient serum, and the specific operation steps are as follows:
[0080] Step 1. According to the instructions, use the TIANamp Viral Genomic DNA Extraction Kit (Tiangen Biochemical Technology (Beijing) Co., Ltd., DP315) to extract HBV genomic DNA from serum samples of hepatitis B patients.
[0081] Step 2, using the first primer pair shown in SEQ ID NO: 1 and SEQ ID NO: 2 to amplify the sample. The PCR amplification system of step 2 is shown in Table 4.
[0082] Table 4 PCR amplification system of step 2
[0083] Element
[0084] The reaction conditions of PCR in step 2 were: pre-denaturation at 95°C for 10 min; denaturation at 95°C for 10 s, annealing at 58°C for 30 s, extension at 72°C for 1 min, a total of 35 cycles of amplification; and final extension at 72°C for 5 min. Thus, a first amplification product is obtained.
[0085] Step 3, respectively using the second prim...
Embodiment 2
[0116] In this embodiment, two kinds of amplification schemes are used to detect the standard substance of gradient dilution. The standard substance is selected from the site-directed mutagenesis artificial plasmid containing the sequence to be detected. The specific operation steps are as follows:
[0117] Option One:
[0118] 1, against 10 8 IU / ml standard was serially diluted to 10 7 IU / ml, 10 6 IU / ml, 10 5 IU / ml, 10 4 IU / ml, 10 3 IU / ml, 10 2 IU / ml, 10IU / ml.
[0119] 2. Using the operation steps of Example 1 to detect the standard substance after the gradient dilution, the results are shown in Table 11.
[0120] The analysis result of the mutation situation of each mutation site of the measured fragment in Table 11
[0121]
[0122] The results show that the operation steps of Scheme 1 can detect the concentration of 10 2 IU / ml of sample.
[0123] Conclusion: This method can effectively detect HBV nucleic acid samples from hepatitis B patients, and the detectab...
Embodiment 3
[0138] In this example, two schemes are used to detect the standard substance of gradient dilution. The standard substance is selected from the site-directed mutagenesis artificial plasmid containing the sequence to be detected. The specific operation steps are as follows:
[0139] Option One:
[0140] 1, against 10 8 IU / ml standard was serially diluted to 10 7 IU / ml, 10 6 IU / ml, 10 5 IU / ml, 10 4 IU / ml, 10 3 IU / ml, 10 2 IU / ml, 10IU / ml.
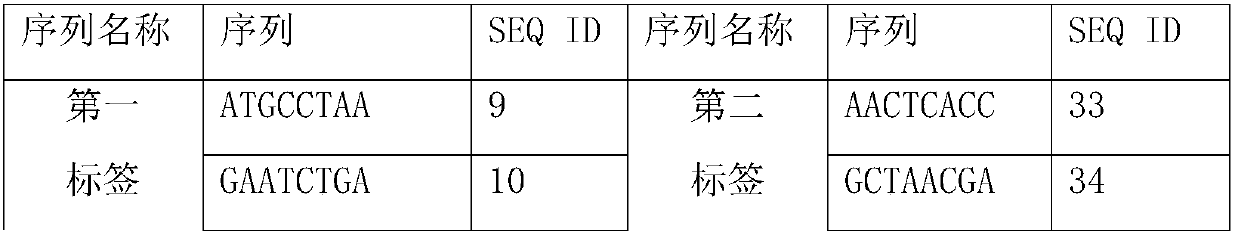
[0141] 2. Using the operation steps of Example 1 to detect the above-mentioned standard substance after gradient dilution, wherein, during the PCR amplification in step 3, multiple drug-resistance-related mutations contained in the above-mentioned two fragments were detected for 8 samples Point, using the tag sequence to distinguish samples, therefore, the third primer and the fifth primer have 8 tag sequences, these 8 tag sequences are shown in SEQ ID NO: 9 - SEQ ID NO: 16, the fourth primer, the sixth primer The primer has a tag sequ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com