Method for detecting human 8-hydroxyl guanine DNA glycosylase activity through circular signal amplification based on autocatalytic replication mediation
A hydroxyguanine, glycosylase biological technology, applied in the field of biological analysis, can solve problems such as limiting wide application, and achieve the effects of reducing experimental background, high sensitivity and low background signal
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
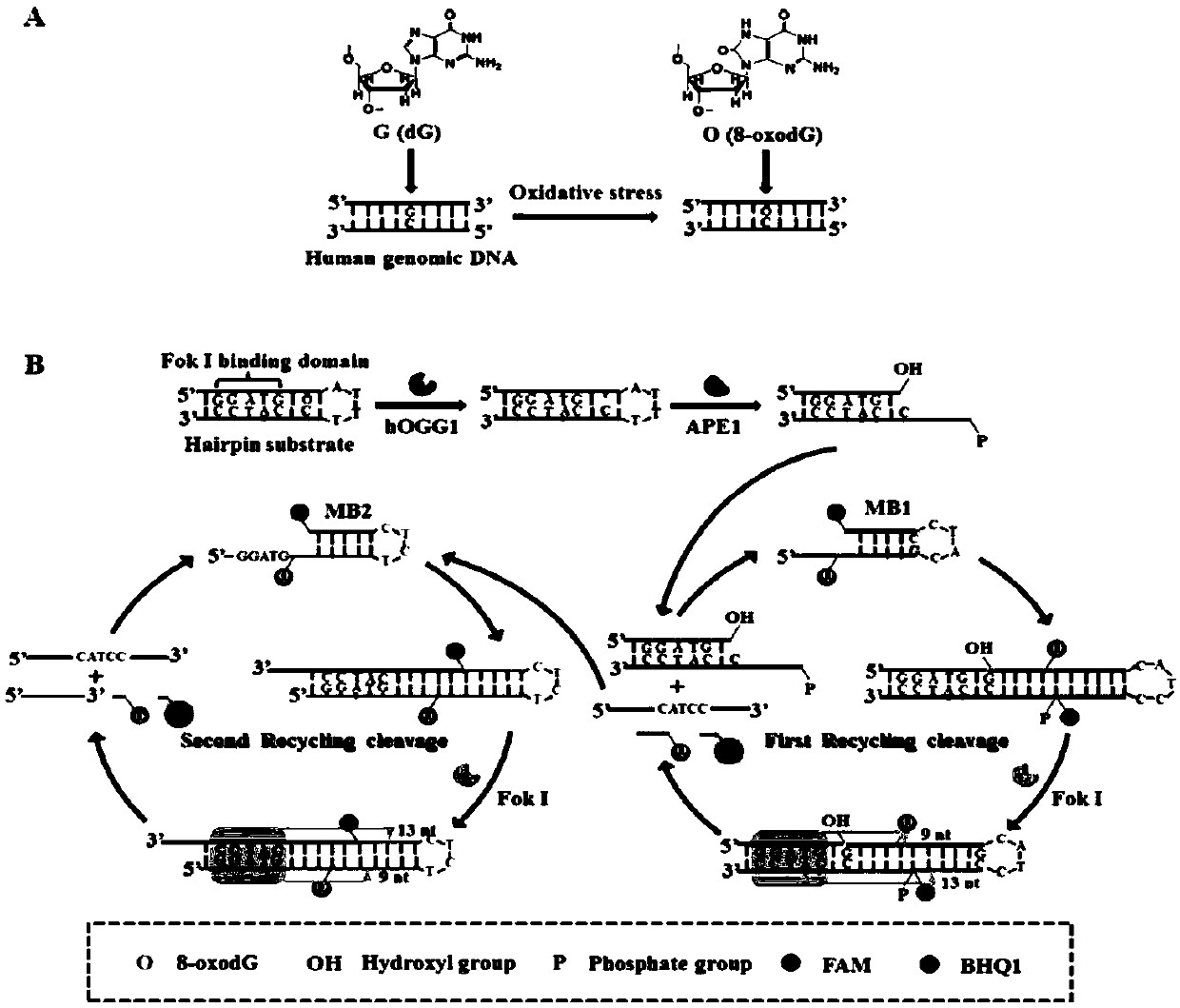
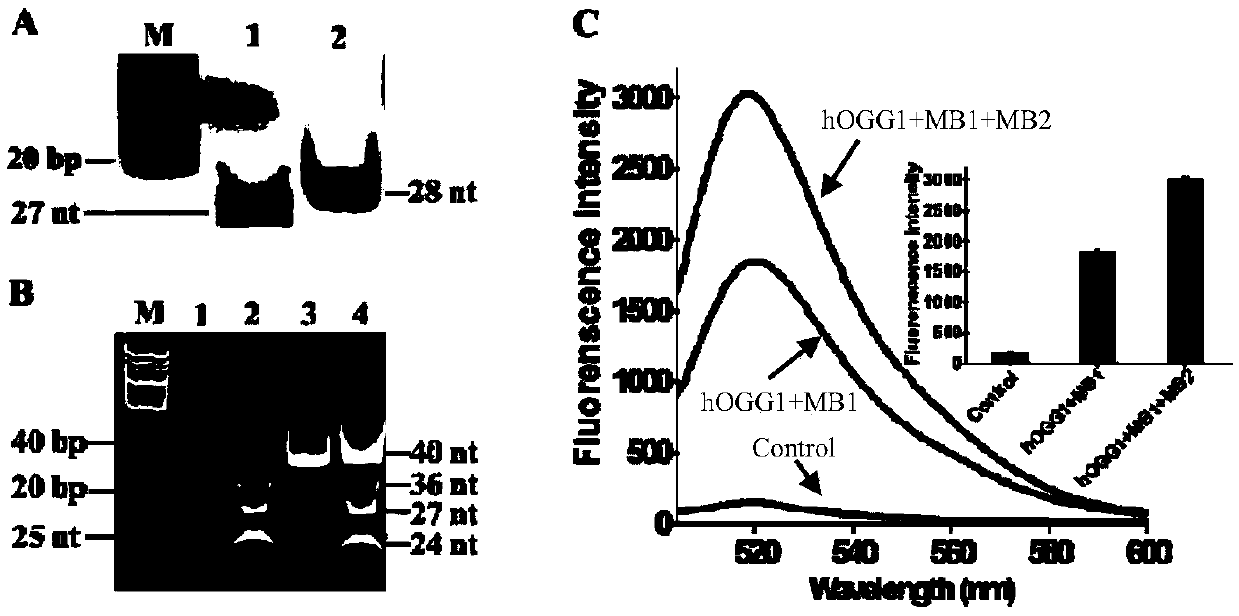
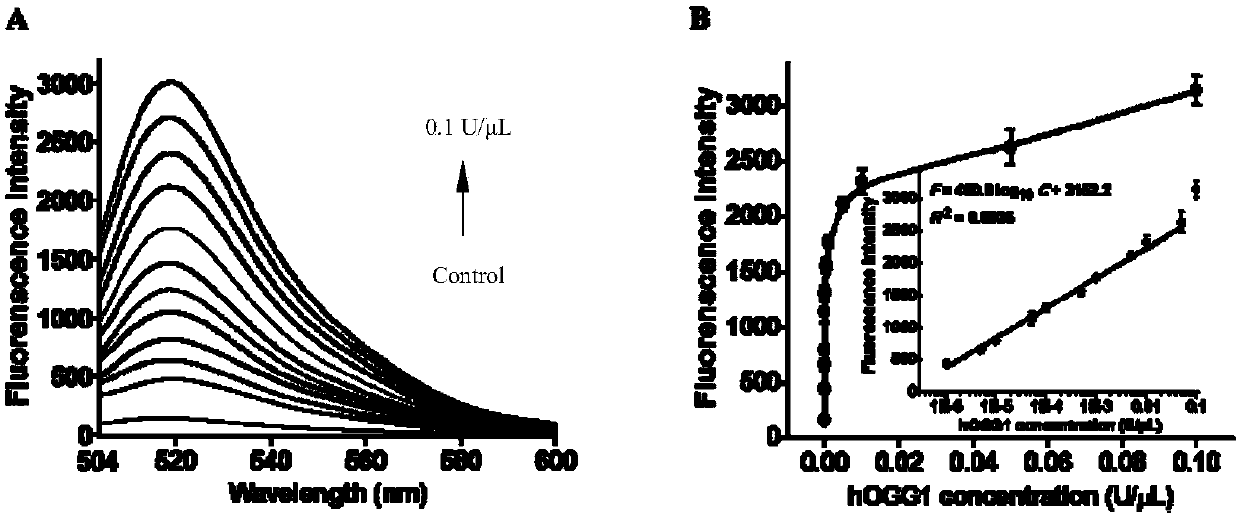
[0062] A method for the ultrasensitive detection of human 8-oxoguanine DNA glycosylase activity based on autocatalytic replication-mediated cyclic signal amplification. The method consists of two sequential reactions: (1) hOGG1-triggered cleavage of the hairpin substrate, and (2) cyclic signal amplification triggered by autocatalytic replication-mediated cascade cleavage of molecular beacons (i.e., MB1 and MB2).
[0063] In the first step, when hOGG1 is present, it will specifically recognize and excise 8-oxoG by cleaving the N-glycosidic bond between the deoxypentose sugar and the damaged base, generating an apurinic / apyrimidinic (AP) site point. The AP site is then efficiently cleaved with the assistance of human apurinic / apyrimidinic endonuclease (APE1), creating a single-nucleotide gap that leads to the unfolding of the loop of the hairpin substrate to form a protruding 9-base DNA sequence.
[0064] In the second step, the unfolded loop sequence (i.e., the 9-nt DNA seque...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com