Gene library construction method of hereditary gynecologic tumors and kit
A technology for gynecological tumors and gene libraries, which is applied in chemical libraries, biochemical equipment and methods, and microbial measurement/testing, and can solve the problems of insufficient DNA and complicated processes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
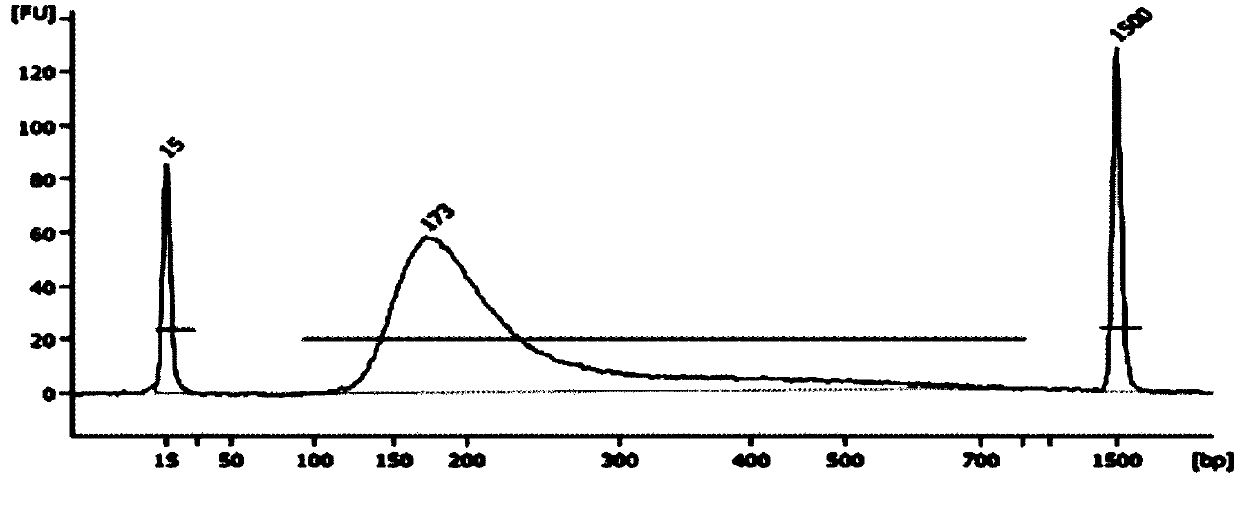
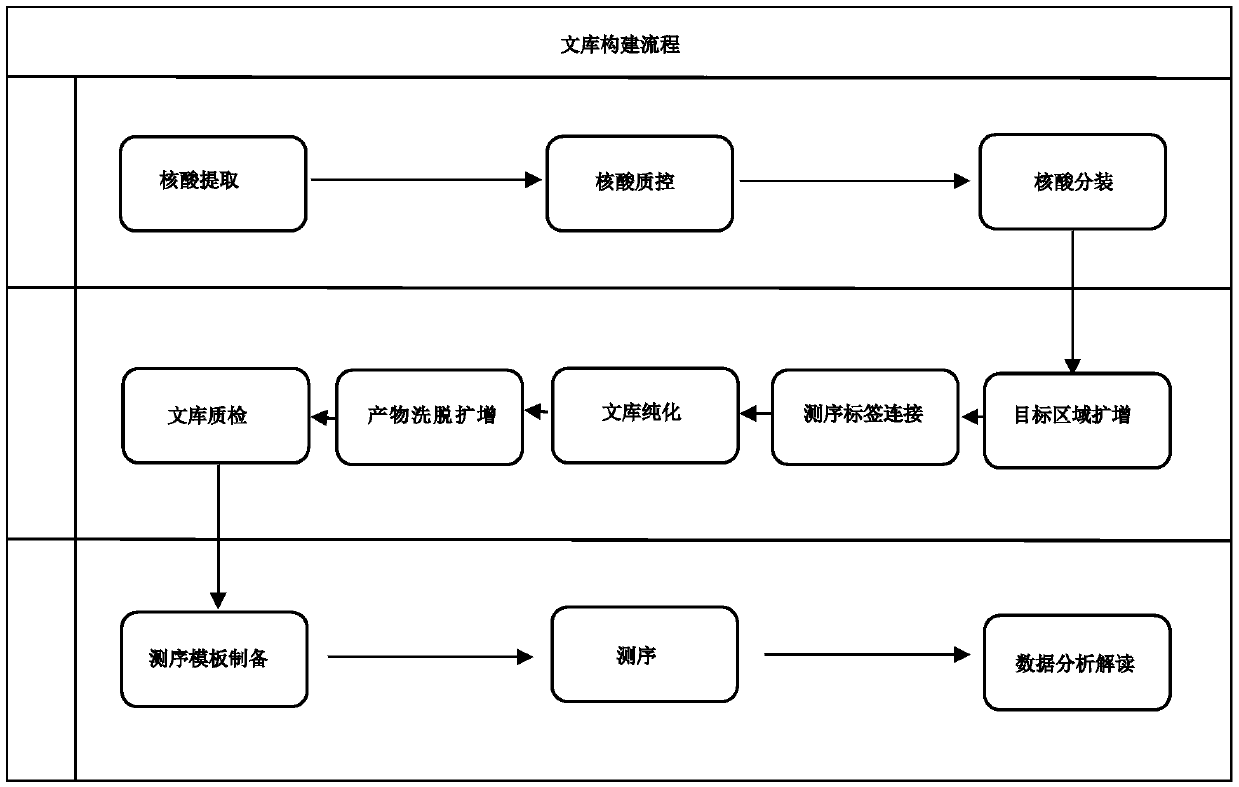
[0108] Genomic DNA extracted from 20 peripheral blood samples was used for library construction, using ATM, BARD1, BRCA1, BRCA2, BRIP1, CDH1, CHEK2, NBN, PIK3CA, PALB2, PTEN, RAD51C, RAD51D, STK11, TP53, MSH2, MLH1, MSH6, PMS2, EPCAM, a total of 20 genes encoding amino acid exon regions and 20 base regions upstream and downstream of the exons are used as the primer set of the target region for the amplification reaction of the target region, combined with the high-throughput sequencing platform IonGeneStudio S5 Sequencing is performed, and single base mutation (SNP) and small fragment insertion-deletion (InDel) detection and analysis are performed on the samples to be tested. The specific operation process is as follows:
[0109] (3) Nucleic acid extraction and quality inspection: Genomic DNA extracted from peripheral blood samples is required to meet certain quality control standards after quality inspection: DNA concentration: ≥1ng / μL; DNA purity: OD260 / 280 1.8-2.0, OD260 / 23...
Embodiment 2
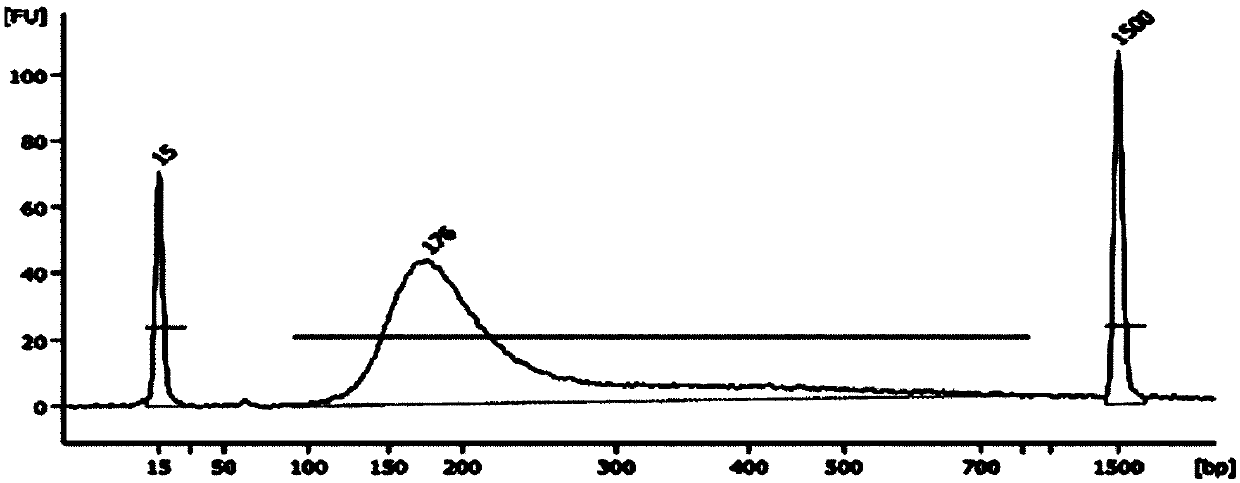
[0122] Genomic DNA extracted from 20 formalin-fixed paraffin-embedded samples was used for library construction, including ATM, BARD1, BRCA1, BRCA2, BRIP1, CDH1, CHEK2, NBN, PIK3CA, PALB2, PTEN, RAD51C, RAD51D, STK11, TP53, MSH2, MLH1, MSH6, PMS2, EPCAM, a total of 20 genes encoding amino acid exon regions and 20 base regions upstream and downstream of the exons are used as the primer set for the target region to amplify the target region, and combined with high The throughput sequencing platform Ion Torrent PGM performs sequencing, and performs single base mutation (SNP) and small fragment insertion-deletion (InDel) detection and analysis on the samples to be tested. The specific operation process is as follows:
[0123] (1) Nucleic acid extraction and quality inspection: Genomic DNA extracted from peripheral blood samples is required to meet certain quality control standards after quality inspection: DNA concentration: ≥1ng / μL; DNA purity: OD260 / 280 1.8-2.0, OD260 / 230 >2; D...
Embodiment 3
[0139] Genomic DNA extracted from 20 buccal swab samples was used for library construction, including ATM, BARD1, BRCA1, BRCA2, BRIP1, CDH1, CHEK2, NBN, PIK3CA, PALB2, PTEN, RAD51C, RAD51D, STK11, TP53, MSH2, MLH1 , MSH6, PMS2, EPCAM, a total of 20 gene coding amino acid exon regions and 20 base regions upstream and downstream of the exons are used as primer sets for the target region to amplify the target region, and combined with the high-throughput sequencing platform IonTorrent PGM is used for sequencing, and single base mutation (SNP) and small fragment insertion-deletion (InDel) detection and analysis are performed on the samples to be tested. The specific operation process is as follows:
[0140] (3) Nucleic acid extraction and quality inspection: Genomic DNA extracted from peripheral blood samples is required to meet certain quality control standards after quality inspection: DNA concentration: ≥1ng / μL; DNA purity: OD260 / 280 1.8-2.0, OD260 / 230 >2; DNA total input amou...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com